Essential Trunk Case
https://www.saksfifthavenue.com/product/rimowa-essential-trunk-case-0400099828331.html?dwvar_0400099828331_color=MATTE%20BLACK




https://www.saksfifthavenue.com/product/rimowa-essential-trunk-case-0400099828331.html?dwvar_0400099828331_color=MATTE%20BLACK
Reconstructive Surgery
Rebodiment
Beauty Surgery
Artificial Organs
Storage for Organs and Shell
Full Rebuild
------------------------------------------------
6'0 30 lb female
10'0 ~100lb male
------------------------------------------------
Odd-toed ungulates, mammals which constitute the taxonomic order Perissodactyla (/pəˌrɪsoʊˈdæktɪlə/, from Ancient Greek περισσός, perissós 'odd', and δάκτυλος, dáktylos 'finger, toe'[3]), are animals—ungulates—who have reduced the weight-bearing toes to three (rhinoceroses and tapirs, with tapirs still using four toes on the front legs) or one (equines, third toe) of the five original toes. The non-weight-bearing toes are either present, absent, vestigial, or positioned posteriorly. By contrast, the even-toed ungulates bear most of their weight equally on four or two (an even number) of the five toes: their third and fourth toes. Another difference between the two is that odd-toed ungulates digest plant cellulose in their intestines rather than in one or more stomach chambers as even-toed ungulates, with the exception of Suina, do.
The order includes about 17 species divided into three families: Equidae (horses, asses, and zebras), Rhinocerotidae (rhinoceroses), and Tapiridae (tapirs).
Despite their very different appearances, they were recognized as related families in the 19th century by the zoologist Richard Owen, who also coined the order name.
https://en.wikipedia.org/wiki/Odd-toed_ungulate
| Eohippus | |
|---|---|

| |
| Reconstructed skeleton, National Museum of Natural History, Washington, DC, United States | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Chordata |
| Class: | Mammalia |
| Order: | Perissodactyla |
| Family: | Equidae |
| Genus: | †Eohippus Marsh, 1876 |
| Species: | †E. angustidens
|
| Binomial name | |
| †Eohippus angustidens (Cope, 1875)
| |
| Synonyms | |
| |
https://en.wikipedia.org/wiki/Eohippus
https://en.wikipedia.org/wiki/Mesohippus
https://en.wikipedia.org/wiki/Protohippus
https://en.wikipedia.org/wiki/Acritohippus
https://en.wikipedia.org/wiki/Archaeohippus
https://en.wikipedia.org/wiki/Dinohippus
https://en.wikipedia.org/wiki/Circulatory_system_of_the_horse
https://en.wikipedia.org/wiki/Monocular_vision
https://en.wikipedia.org/wiki/Chestnut_(horse_anatomy)
https://en.wikipedia.org/wiki/Hinny
https://en.wikipedia.org/wiki/Genomic_imprinting
https://en.wikipedia.org/wiki/Uniparental_disomy
https://en.wikipedia.org/wiki/Trisomic_rescue
https://en.wikipedia.org/wiki/Isodisomy
Uniparental disomy (UPD) occurs when a person receives two copies of a chromosome, or of part of a chromosome, from one parent and no copy from the other parent.[1] UPD can be the result of heterodisomy, in which a pair of non-identical chromosomes are inherited from one parent (an earlier stage meiosis I error) or isodisomy, in which a single chromosome from one parent is duplicated (a later stage meiosis II error).[2] Uniparental disomy may have clinical relevance for several reasons. For example, either isodisomy or heterodisomy can disrupt parent-specific genomic imprinting, resulting in imprinting disorders. Additionally, isodisomy leads to large blocks of homozygosity, which may lead to the uncovering of recessive genes, a similar phenomenon seen in inbred children of consanguineous partners.[3]
UPD has been found to occur in about 1 in 2,000 births.[4]
UPD can occur as a random event during the formation of egg cells or sperm cells or may happen in early fetal development. It can also occur during trisomic rescue.
Most occurrences of UPD result in no phenotypical anomalies. However, if the UPD-causing event happened during meiosis II, the genotype may include identical copies of the uniparental chromosome (isodisomy), leading to the manifestation of rare recessive disorders. UPD should be suspected in an individual manifesting a recessive disorder where only one parent is a carrier.
Uniparental inheritance of imprinted genes can also result in phenotypical anomalies. Although few imprinted genes have been identified, uniparental inheritance of an imprinted gene can result in the loss of gene function, which can lead to delayed development, intellectual disability, or other medical problems.[citation needed]
UPD has rarely been studied prospectively, with most reports focusing on either known conditions or incidental findings. It has been proposed that the incidence may not be as low as believed, rather it may be under-reported.[9]
Genome wide UPD, also called uniparental diploidy, is when all chromosomes are inherited from one parent. Only in mosaic form can this phenomenon be compatible with life. As of 2017, there have only been 18 reported cases of genome wide UPD.[10]
The first clinical case of UPD was reported in 1988 and involved a girl with cystic fibrosis and short stature who carried two copies of maternal chromosome 7.[11] Since 1991, out of the 47 possible disomies, 29 have been identified among individuals ascertained for medical reasons. This includes chromosomes 2, 5–11, 13–16, 21 and 22.
https://en.wikipedia.org/wiki/Uniparental_disomy
https://en.wikipedia.org/wiki/Chromosome_14
https://en.wikipedia.org/wiki/ACIN1
https://en.wikipedia.org/wiki/FSCB
https://en.wikipedia.org/wiki/Monosomy_14
https://en.wikipedia.org/wiki/Nonsyndromic_deafness
https://en.wikipedia.org/wiki/Chromosome
| Chromosome 19 | |
|---|---|
 Human chromosome 19 pair after G-banding. One is from mother, one is from father. | |
 Chromosome 19 pair in human male karyogram. | |
| Features | |
| Length (bp) | 61,707,364 bp (CHM13) |
| No. of genes | 1,357 (CCDS)[1] |
| Type | Autosome |
| Centromere position | Metacentric[2] (26.2 Mbp[3]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome 19 |
| Entrez | Chromosome 19 |
| NCBI | Chromosome 19 |
| UCSC | Chromosome 19 |
| Full DNA sequences | |
| RefSeq | NC_000019 (FASTA) |
| GenBank | CM000681 (FASTA) |
Chromosome 19 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 19 spans more than 61.7 million base pairs, the building material of DNA. It is considered the most gene-rich chromosome containing roughly 1,500 genes, despite accounting for only 2 percent of the human genome.[4][5]
The following are some of the gene count estimates of human chromosome 19. Because researchers use different approaches to genome annotation, their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[6]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 1,357 | — | — | [1] | 2016-09-08 |
| HGNC | 1,372 | 299 | 413 | [7] | 2017-05-12 |
| Ensembl | 1,469 | 894 | 514 | [8] | 2017-03-29 |
| UniProt | 1,435 | — | — | [9] | 2018-02-28 |
| NCBI | 1,430 | 604 | 528 | [10][11][12] | 2017-05-19 |
The following is a partial list of genes on human chromosome 19. For complete list, see the link in the infobox on the right.
The following diseases are some of those related to genes on chromosome 19:[15]
| Chr. | Arm[23] | Band[24] | ISCN start[25] |
ISCN stop[25] |
Basepair start |
Basepair stop |
Stain[26] | Density |
|---|---|---|---|---|---|---|---|---|
| 19 | p | 13.3 | 0 | 578 | 1 | 6,900,000 | gneg |
|
| 19 | p | 13.2 | 578 | 870 | 6,900,001 | 12,600,000 | gpos | 25 |
| 19 | p | 13.13 | 870 | 1034 | 12,600,001 | 13,800,000 | gneg |
|
| 19 | p | 13.12 | 1034 | 1216 | 13,800,001 | 16,100,000 | gpos | 25 |
| 19 | p | 13.11 | 1216 | 1581 | 16,100,001 | 19,900,000 | gneg |
|
| 19 | p | 12 | 1581 | 1809 | 19,900,001 | 24,200,000 | gvar |
|
| 19 | p | 11 | 1809 | 1992 | 24,200,001 | 26,200,000 | acen |
|
| 19 | q | 11 | 1992 | 2159 | 26,200,001 | 28,100,000 | acen |
|
| 19 | q | 12 | 2159 | 2372 | 28,100,001 | 31,900,000 | gvar |
|
| 19 | q | 13.11 | 2372 | 2569 | 31,900,001 | 35,100,000 | gneg |
|
| 19 | q | 13.12 | 2569 | 2737 | 35,100,001 | 37,800,000 | gpos | 25 |
| 19 | q | 13.13 | 2737 | 2949 | 37,800,001 | 38,200,000 | gneg |
|
| 19 | q | 13.2 | 2949 | 3101 | 38,200,001 | 42,900,000 | gpos | 25 |
| 19 | q | 13.31 | 3101 | 3193 | 42,900,001 | 44,700,000 | gneg |
|
| 19 | q | 13.32 | 3193 | 3390 | 44,700,001 | 47,500,000 | gpos | 25 |
| 19 | q | 13.33 | 3390 | 3649 | 47,500,001 | 50,900,000 | gneg |
|
| 19 | q | 13.41 | 3649 | 3770 | 50,900,001 | 53,100,000 | gpos | 25 |
| 19 | q | 13.42 | 3770 | 3938 | 53,100,001 | 55,800,000 | gneg |
|
| 19 | q | 13.43 | 3938 | 4120 | 55,800,001 | 58,617,616 | gpos | 25 |
https://en.wikipedia.org/wiki/Chromosome_19
The following are some of the gene count estimates of human chromosome 10. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[4]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 706 | — | — | [1] | 2016-09-08 |
| HGNC | 708 | 244 | 614 | [5] | 2017-05-12 |
| Ensembl | 728 | 881 | 568 | [6] | 2017-03-29 |
| UniProt | 750 | — | — | [7] | 2018-02-28 |
| NCBI | 754 | 842 | 654 | [8][9][10] | 2017-05-19 |
The following is a partial list of genes on human chromosome 10. For complete list, see the link in the infobox on the right.
The following diseases are related to genes on chromosome 10:
| Chr. | Arm[16] | Band[17] | ISCN start[18] |
ISCN stop[18] |
Basepair start |
Basepair stop |
Stain[19] | Density |
|---|---|---|---|---|---|---|---|---|
| 10 | p | 15.3 | 0 | 229 | 1 | 3,000,000 | gneg |
|
| 10 | p | 15.2 | 229 | 329 | 3,000,001 | 3,800,000 | gpos | 25 |
| 10 | p | 15.1 | 329 | 630 | 3,800,001 | 6,600,000 | gneg |
|
| 10 | p | 14 | 630 | 917 | 6,600,001 | 12,200,000 | gpos | 75 |
| 10 | p | 13 | 917 | 1175 | 12,200,001 | 17,300,000 | gneg |
|
| 10 | p | 12.33 | 1175 | 1361 | 17,300,001 | 18,300,000 | gpos | 75 |
| 10 | p | 12.32 | 1361 | 1432 | 18,300,001 | 18,400,000 | gneg |
|
| 10 | p | 12.31 | 1432 | 1604 | 18,400,001 | 22,300,000 | gpos | 75 |
| 10 | p | 12.2 | 1604 | 1662 | 22,300,001 | 24,300,000 | gneg |
|
| 10 | p | 12.1 | 1662 | 1891 | 24,300,001 | 29,300,000 | gpos | 50 |
| 10 | p | 11.23 | 1891 | 2063 | 29,300,001 | 31,100,000 | gneg |
|
| 10 | p | 11.22 | 2063 | 2235 | 31,100,001 | 34,200,000 | gpos | 25 |
| 10 | p | 11.21 | 2235 | 2406 | 34,200,001 | 38,000,000 | gneg |
|
| 10 | p | 11.1 | 2406 | 2621 | 38,000,001 | 39,800,000 | acen |
|
| 10 | q | 11.1 | 2621 | 2850 | 39,800,001 | 41,600,000 | acen |
|
| 10 | q | 11.21 | 2850 | 3051 | 41,600,001 | 45,500,000 | gneg |
|
| 10 | q | 11.22 | 3051 | 3252 | 45,500,001 | 48,600,000 | gpos | 25 |
| 10 | q | 11.23 | 3252 | 3409 | 48,600,001 | 51,100,000 | gneg |
|
| 10 | q | 21.1 | 3409 | 3753 | 51,100,001 | 59,400,000 | gpos | 100 |
| 10 | q | 21.2 | 3753 | 3839 | 59,400,001 | 62,800,000 | gneg |
|
| 10 | q | 21.3 | 3839 | 4097 | 62,800,001 | 68,800,000 | gpos | 100 |
| 10 | q | 22.1 | 4097 | 4469 | 68,800,001 | 73,100,000 | gneg |
|
| 10 | q | 22.2 | 4469 | 4655 | 73,100,001 | 75,900,000 | gpos | 50 |
| 10 | q | 22.3 | 4655 | 4970 | 75,900,001 | 80,300,000 | gneg |
|
| 10 | q | 23.1 | 4970 | 5200 | 80,300,001 | 86,100,000 | gpos | 100 |
| 10 | q | 23.2 | 5200 | 5331 | 86,100,001 | 87,700,000 | gneg |
|
| 10 | q | 23.31 | 5331 | 5558 | 87,700,001 | 91,100,000 | gpos | 75 |
| 10 | q | 23.32 | 5558 | 5672 | 91,100,001 | 92,300,000 | gneg |
|
| 10 | q | 23.33 | 5672 | 5887 | 92,300,001 | 95,300,000 | gpos | 50 |
| 10 | q | 24.1 | 5887 | 5973 | 95,300,001 | 97,500,000 | gneg |
|
| 10 | q | 24.2 | 5973 | 6131 | 97,500,001 | 100,100,000 | gpos | 50 |
| 10 | q | 24.31 | 6131 | 6202 | 100,100,001 | 101,200,000 | gneg |
|
| 10 | q | 24.32 | 6202 | 6317 | 101,200,001 | 103,100,000 | gpos | 25 |
| 10 | q | 24.33 | 6317 | 6374 | 103,100,001 | 104,000,000 | gneg |
|
| 10 | q | 25.1 | 6374 | 6646 | 104,000,001 | 110,100,000 | gpos | 100 |
| 10 | q | 25.2 | 6646 | 6761 | 110,100,001 | 113,100,000 | gneg |
|
| 10 | q | 25.3 | 6761 | 6890 | 113,100,001 | 117,300,000 | gpos | 75 |
| 10 | q | 26.11 | 6890 | 7090 | 117,300,001 | 119,900,000 | gneg |
|
| 10 | q | 26.12 | 7090 | 7219 | 119,900,001 | 121,400,000 | gpos | 50 |
| 10 | q | 26.13 | 7219 | 7506 | 121,400,001 | 125,700,000 | gneg |
|
| 10 | q | 26.2 | 7506 | 7721 | 125,700,001 | 128,800,000 | gpos | 50 |
| 10 | q | 26.3 | 7721 | 8050 | 128,800,001 | 133,797,422 | gneg |
|
https://en.wikipedia.org/wiki/Chromosome_10
https://en.wikipedia.org/wiki/Chromosome_9
https://en.wikipedia.org/wiki/Marfan_syndrome
https://en.wikipedia.org/wiki/Multiple_epiphyseal_dysplasia
https://en.wikipedia.org/wiki/Centronuclear_myopathy
https://en.wikipedia.org/wiki/Spinocerebellar_ataxia_type_6
https://en.wikipedia.org/wiki/Uniparental_disomy
https://en.wikipedia.org/wiki/Platelet-derived_growth_factor
https://en.wikipedia.org/wiki/Chromosomal_translocation
https://en.wikipedia.org/wiki/Resection_margin
https://en.wikipedia.org/wiki/Magnetic_resonance_imaging
https://en.wikipedia.org/wiki/TAR_syndrome
https://en.wikipedia.org/wiki/Uniparental_disomy
https://en.wikipedia.org/wiki/Dermatofibrosarcoma_protuberans
https://en.wikipedia.org/wiki/Trisomy_X
https://en.wikipedia.org/wiki/Fragile_X_syndrome
https://en.wikipedia.org/wiki/Acute_megakaryoblastic_leukemia
https://en.wikipedia.org/wiki/Acute_promyelocytic_leukemia
https://en.wikipedia.org/wiki/Myxoid_liposarcoma
https://en.wikipedia.org/wiki/Alveolar_rhabdomyosarcoma
https://en.wikipedia.org/wiki/Category:Neurogenetic_disorders
https://en.wikipedia.org/wiki/Category:Syndromes_with_craniofacial_abnormalities
https://en.wikipedia.org/wiki/Category:Rare_syndromes
https://en.wikipedia.org/wiki/Category:Syndromes_affecting_the_eye
https://en.wikipedia.org/wiki/Category:Trinucleotide_repeat_disorders
https://en.wikipedia.org/wiki/Deaflympics
https://en.wikipedia.org/wiki/Alien_hand_syndrome
https://en.wikipedia.org/wiki/Antisynthetase_syndrome
https://en.wikipedia.org/wiki/ATR-16_syndrome
https://en.wikipedia.org/wiki/Birt%E2%80%93Hogg%E2%80%93Dub%C3%A9_syndrome
https://en.wikipedia.org/wiki/Blue_diaper_syndrome
https://en.wikipedia.org/wiki/Fraser_syndrome
https://en.wikipedia.org/wiki/Felty%27s_syndrome
https://en.wikipedia.org/wiki/Gray_platelet_syndrome
https://en.wikipedia.org/wiki/Greig_cephalopolysyndactyly_syndrome
https://en.wikipedia.org/wiki/Greig_cephalopolysyndactyly_syndrome
https://en.wikipedia.org/wiki/Hereditary_fibrosing_poikiloderma_with_tendon_contractures,_myopathy,_and_pulmonary_fibrosis
https://en.wikipedia.org/wiki/Immunodeficiency_26
https://en.wikipedia.org/wiki/Joubert_syndrome
https://en.wikipedia.org/wiki/Hyperphosphatasia_with_mental_retardation_syndrome
https://en.wikipedia.org/wiki/Jalili_syndrome
https://en.wikipedia.org/wiki/Kaufman_oculocerebrofacial_syndrome
https://en.wikipedia.org/wiki/Keratitis%E2%80%93ichthyosis%E2%80%93deafness_syndrome
https://en.wikipedia.org/wiki/KBG_syndrome
https://en.wikipedia.org/wiki/13q_deletion_syndrome
https://en.wikipedia.org/wiki/De_novo_mutation
https://en.wikipedia.org/wiki/Syndactyly-nystagmus_syndrome_due_to_2q31.1_microduplication
https://en.wikipedia.org/wiki/Beare%E2%80%93Stevenson_cutis_gyrata_syndrome
https://en.wikipedia.org/wiki/Carpenter_syndrome
https://en.wikipedia.org/wiki/CLOVES_syndrome
https://en.wikipedia.org/wiki/COACH_syndrome
https://en.wikipedia.org/wiki/Marshall_syndrome
https://en.wikipedia.org/wiki/L1_syndrome
https://en.wikipedia.org/wiki/IVIC_syndrome
https://en.wikipedia.org/wiki/Graham%E2%80%93Boyle%E2%80%93Troxell_syndrome
https://en.wikipedia.org/wiki/Fibular_aplasia-ectrodactyly_syndrome
https://en.wikipedia.org/wiki/Microcephaly_albinism_digital_anomalies_syndrome
https://en.wikipedia.org/wiki/Skin_fragility-woolly_hair-palmoplantar_keratoderma_syndrome
https://en.wikipedia.org/wiki/Syndactyly-nystagmus_syndrome_due_to_2q31.1_microduplication
| Syndactyly-nystagmus syndrome due to 2q31.1 duplication | |
|---|---|
| Other names | 2q31.1 microduplication syndrome |
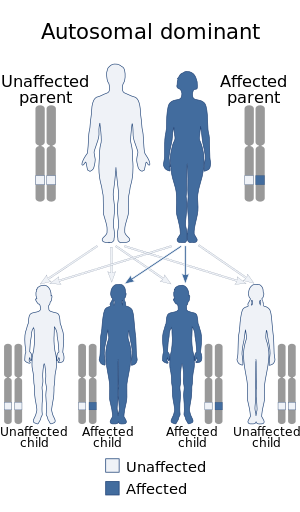 | |
| The microduplication associated with this condition is autosomal dominant | |
| Specialty | Medical genetics |
| Symptoms | Mainly syndactyly and congenital bilateral pendular nystagmus |
| Complications | none |
| Usual onset | birth |
| Duration | lifelong (unless surgically corrected) |
| Causes | genetic mutation (more specifically an autosomal dominant chromosomal microduplication containing HOX genes) |
| Prevention | none |
| Prognosis | good |
| Frequency | rare |
| Deaths | - |
Syndactyly-nystagmus syndrome due to 2q31.1 microduplication, also known as 2q31.1 microduplication syndrome, is a rare genetic disorder characterized by syndactyly affecting the third-fourth fingers and bilateral congenital nystagmus.[1]
https://en.wikipedia.org/wiki/Syndactyly-nystagmus_syndrome_due_to_2q31.1_microduplication
https://en.wikipedia.org/wiki/Triphalangeal_thumb
https://en.wikipedia.org/wiki/Chromosome_2
https://en.wikipedia.org/wiki/Gene_duplication
https://en.wikipedia.org/wiki/Homeobox
https://en.wikipedia.org/wiki/Fungus
https://en.wikipedia.org/wiki/Switzerland
https://en.wikipedia.org/wiki/Bilateria
https://en.wikipedia.org/wiki/Antiparallel_(biochemistry)
https://en.wikipedia.org/wiki/Homeobox
https://en.wikipedia.org/wiki/Lambda_phage
https://en.wikipedia.org/wiki/Angiogenesis
https://en.wikipedia.org/wiki/Gene_duplication
https://en.wikipedia.org/wiki/Unequal_crossing-over
https://en.wikipedia.org/wiki/Gene_conversion
https://en.wikipedia.org/wiki/Concerted_evolution
https://en.wikipedia.org/wiki/Pseudoautosomal_region
https://en.wikipedia.org/wiki/Pseudogene
https://en.wikipedia.org/wiki/Gene_conversion
https://en.wikipedia.org/wiki/Gene_duplication
https://en.wikipedia.org/wiki/Category:Modification_of_genetic_information
https://en.wikipedia.org/wiki/Bacterial_conjugation
https://en.wikipedia.org/wiki/Fusion_gene
https://en.wikipedia.org/wiki/Transfection
https://en.wikipedia.org/wiki/Antigenic_shift
https://en.wikipedia.org/wiki/Macrosatellite
https://en.wikipedia.org/wiki/Retroposon
https://en.wikipedia.org/wiki/LINE1
https://en.wikipedia.org/wiki/P_element
https://en.wikipedia.org/wiki/Gene_amplification
https://en.wikipedia.org/wiki/Gene_cluster
https://en.wikipedia.org/wiki/Pathogenicity_island
https://en.wikipedia.org/wiki/Low_copy_repeats
https://en.wikipedia.org/wiki/Directional_selection
https://en.wikipedia.org/wiki/Negative_selection_(natural_selection)
https://en.wikipedia.org/wiki/Stabilizing_selection
https://en.wikipedia.org/wiki/Balancing_selection
https://en.wikipedia.org/wiki/Synonymous_substitution
https://en.wikipedia.org/wiki/Silent_mutation
https://en.wikipedia.org/wiki/Nonsynonymous_substitution
https://en.wikipedia.org/wiki/Stop_codon#Nonstop
https://en.wikipedia.org/wiki/Cytotoxicity
https://en.wikipedia.org/wiki/Stop_codon#Nonstop
https://en.wikipedia.org/wiki/Ribosomal_frameshift
https://en.wikipedia.org/wiki/Mycoplasma_laboratorium#Watermarks
https://en.wikipedia.org/wiki/Genetic_use_restriction_technology
https://en.wikipedia.org/wiki/Chromosome_segregation
https://en.wikipedia.org/wiki/Synthesis-dependent_strand_annealing
https://en.wikipedia.org/wiki/DNA_repair#Double-strand_breaks
https://en.wikipedia.org/wiki/V(D)J_recombination
https://en.wikipedia.org/wiki/DNA_end_resection
https://en.wikipedia.org/wiki/Pyrimidine_dimer#Mutagenesis
https://en.wikipedia.org/wiki/Lesion
https://en.wikipedia.org/wiki/Osteolytic_lesion
https://en.wikipedia.org/wiki/Osteoporosis_circumscripta
https://en.wikipedia.org/wiki/Ghon_focus
https://en.wikipedia.org/wiki/Ionizing_radiation
https://en.wikipedia.org/wiki/DNA_repair#Double-strand_breaks
https://en.wikipedia.org/wiki/Pyrimidine_dimer
https://en.wikipedia.org/wiki/ATM_serine/threonine_kinase
https://en.wikipedia.org/wiki/Signal_transduction
https://en.wikipedia.org/wiki/Signal_transduction
https://en.wikipedia.org/wiki/Repressor
https://en.wikipedia.org/wiki/Spirochaete
https://en.wikipedia.org/wiki/Deinococcus_radiodurans
https://en.wikipedia.org/wiki/Calorie_restriction
https://en.wikipedia.org/wiki/Caenorhabditis_elegans
https://en.wikipedia.org/wiki/Gene_dosage
https://en.wikipedia.org/wiki/Trichothiodystrophy
https://en.wikipedia.org/wiki/Ataxia%E2%80%93telangiectasia
https://en.wikipedia.org/wiki/Progeroid_syndromes
https://en.wikipedia.org/wiki/DNA_repair-deficiency_disorder
https://en.wikipedia.org/wiki/Missense_mutation
https://en.wikipedia.org/wiki/File:DNA_damage,_repair,_alteration_of_repair_in_cancer.png
https://en.wikipedia.org/wiki/DNA_repair#translesion_synthesis
https://en.wikipedia.org/wiki/Bacteriophage
https://en.wikipedia.org/wiki/Structural_motif
https://en.wikipedia.org/wiki/Great_Oxidation_Event
https://en.wikipedia.org/wiki/Fossil
https://en.wikipedia.org/wiki/Germline
https://en.wikipedia.org/wiki/Chromosome
https://en.wikipedia.org/wiki/Genetic_diversity
https://en.wikipedia.org/wiki/Mitotic_catastrophe
https://en.wikipedia.org/wiki/Schizosaccharomyces_pombe
https://en.wikipedia.org/wiki/Genetically_modified_crops
https://en.wikipedia.org/wiki/Genome_instability
https://en.wikipedia.org/wiki/Spindle_checkpoint
https://en.wikipedia.org/wiki/Anaphase-promoting_complex
https://en.wikipedia.org/wiki/Kinesin-like_protein_KIF11
https://en.wikipedia.org/wiki/Genetic_engineering
https://en.wikipedia.org/wiki/Recombinant_DNA
https://en.wikipedia.org/wiki/Chromosome
https://en.wikipedia.org/wiki/DNA_microarray
https://en.wikipedia.org/wiki/Operon
https://en.wikipedia.org/wiki/Cistron
https://en.wikipedia.org/wiki/Operon#Operator
https://en.wikipedia.org/wiki/Nematode
https://en.wikipedia.org/wiki/Regulon
https://en.wikipedia.org/wiki/Structural_gene
https://en.wikipedia.org/wiki/Inducer
https://en.wikipedia.org/wiki/Corepressor
https://en.wikipedia.org/wiki/TcoF-DB
https://en.wikipedia.org/wiki/Negative_feedback
https://en.wikipedia.org/wiki/NRIP1
https://en.wikipedia.org/wiki/Acute_myeloid_leukemia
https://en.wikipedia.org/wiki/BCL-6_corepressor
https://en.wikipedia.org/wiki/Rheumatoid_arthritis
https://en.wikipedia.org/wiki/Regulator_gene
https://en.wikipedia.org/wiki/Derepression
https://en.wikipedia.org/wiki/Attenuator_(genetics)
https://en.wikipedia.org/wiki/Genetic_code
https://en.wikipedia.org/wiki/Gene_regulatory_network
https://en.wikipedia.org/wiki/Gal_operon
https://en.wikipedia.org/wiki/Internal_control_region
https://en.wikipedia.org/wiki/Intrinsic_termination
https://en.wikipedia.org/wiki/Neoteny
https://en.wikipedia.org/wiki/Heterochrony
https://en.wikipedia.org/wiki/Neoteny_in_humans
https://en.wikipedia.org/wiki/Fecundity
https://en.wikipedia.org/wiki/Northwestern_salamander
https://en.wikipedia.org/wiki/Centriole
https://en.wikipedia.org/wiki/Microtubule
https://en.wikipedia.org/wiki/Flagellum
https://en.wikipedia.org/wiki/Gram-negative_bacteria
https://en.wikipedia.org/wiki/Rotating_locomotion_in_living_systems
https://en.wikipedia.org/wiki/Type_III_secretion_system
https://en.wikipedia.org/wiki/Chemiosmosis#Proton-motive_force
https://en.wikipedia.org/wiki/Electrochemical_gradient
https://en.wikipedia.org/wiki/Mitochondria
https://en.wikipedia.org/wiki/Molecular_diffusion
https://en.wikipedia.org/wiki/Photophosphorylation
Chemiosmotic phosphorylation is the third pathway that produces ATP from inorganic phosphate and an ADP molecule. This process is part of oxidative phosphorylation.
https://en.wikipedia.org/wiki/Chemiosmosis#Proton-motive_force
https://en.wikipedia.org/wiki/Chemiosmosis#Proton-motive_force
https://en.wikipedia.org/wiki/Ion_channel
https://en.wikipedia.org/wiki/Convection
https://en.wikipedia.org/wiki/Ferredoxin
https://en.wikipedia.org/wiki/Plastoquinone
https://en.wikipedia.org/wiki/Cytochrome_b6f_complex
https://en.wikipedia.org/wiki/Plastocyanin
https://en.wikipedia.org/wiki/Transient_receptor_potential_channel
https://en.wikipedia.org/wiki/Inward-rectifier_potassium_channel
https://en.wikipedia.org/wiki/Metalloprotein
https://en.wikipedia.org/wiki/Iron%E2%80%93sulfur_cluster
https://en.wikipedia.org/wiki/High_potential_iron%E2%80%93sulfur_protein
https://en.wikipedia.org/wiki/Paracoccus_denitrificans