This article needs additional citations for verification. (March 2016) |
 | ||||||||||||||||||||||||||||||||||||||||||||||||
| Greek alphabet | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||
| History | ||||||||||||||||||||||||||||||||||||||||||||||||
| Use in other languages | ||||||||||||||||||||||||||||||||||||||||||||||||
| Related topics | ||||||||||||||||||||||||||||||||||||||||||||||||
Gamma /ˈɡæmə/[1] (uppercase Γ, lowercase γ; Greek: γάμμα gámma) is the third letter of the Greek alphabet. In the system of Greek numerals it has a value of 3. In Ancient Greek, the letter gamma represented a voiced velar stop IPA: [ɡ]. In Modern Greek, this letter represents either a voiced velar fricative IPA: [ɣ] or a voiced palatal fricative IPA: [ʝ] (while /g/ in foreign words is instead commonly transcribed as γκ).
In the International Phonetic Alphabet and other modern Latin-alphabet based phonetic notations, it represents the voiced velar fricative.
History
The Greek letter Gamma Γ is a grapheme derived from the Phoenician letter 𐤂 (gīml) which was rotated from the right-to-left script of Canaanite to accommodate the Greek language's writing system of left-to-right. The Canaanite grapheme represented the /g/ phoneme in the Canaanite language, and as such is cognate with gimel ג of the Hebrew alphabet.
Based on its name, the letter has been interpreted as an abstract representation of a camel's neck,[2] but this has been criticized as contrived,[3] and it is more likely that the letter is derived from an Egyptian hieroglyph representing a club or throwing stick.[4]
In Archaic Greece, the shape of gamma was closer to a classical lambda (Λ), while lambda retained the Phoenician L-shape (𐌋).
Letters that arose from the Greek gamma include Etruscan (Old Italic) 𐌂, Roman C and G, Runic kaunan ᚲ, Gothic geuua 𐌲, the Coptic Ⲅ, and the Cyrillic letters Г and Ґ.[5]
Greek phoneme
The Ancient Greek /g/ phoneme was the voiced velar stop, continuing the reconstructed proto-Indo-European *g, *ǵ.
The modern Greek phoneme represented by gamma is realized either as a voiced palatal fricative (/ʝ/) before a front vowel (/e/, /i/), or as a voiced velar fricative /ɣ/ in all other environments. Both in Ancient and in Modern Greek, before other velar consonants (κ, χ, ξ – that is, k, kh, ks), gamma represents a velar nasal /ŋ/. A double gamma γγ (e.g., άγγελος, "angel") represents the sequence /ŋɡ/ (phonetically varying [ŋɡ~ɡ]) or /ŋɣ/.
Phonetic transcription
Lowercase Greek gamma is used in the Americanist phonetic notation and Uralic Phonetic Alphabet to indicate voiced consonants.
The gamma was also added to the Latin alphabet, as Latin gamma, in the following forms: majuscule Ɣ, minuscule ɣ, and superscript modifier letter ˠ.
In the International Phonetic Alphabet the minuscule letter is used to represent a voiced velar fricative and the superscript modifier letter is used to represent velarization. It is not to be confused with the character ɤ, which looks like a lowercase Latin gamma that lies above the baseline rather than crossing, and which represents the close-mid back unrounded vowel. In certain nonstandard variations of the IPA, the uppercase form is used.[citation needed]
It is as a full-fledged majuscule and minuscule letter in the alphabets of some of languages of Africa such as Dagbani, Dinka, Kabye, and Ewe,[6] and Berber languages using the Berber Latin alphabet.
It is sometimes also used in the romanization of Pashto.
Mathematics and science
Lowercase
The lowercase letter is used as a symbol for:
- Chromatic number of in graph theory
- Gamma radiation in nuclear physics
- The photon, the elementary particle of light and other electromagnetic radiation
- The 434 nm spectral line in the Balmer series
- Surface energy in materials science
- The Lorentz factor in the theory of relativity
- In mathematics, the lower incomplete gamma function
- The Euler–Mascheroni constant ≈ 0.57721566490153286
- The heat capacity ratio Cp /Cv in thermodynamics
- The activity coefficient in thermodynamics
- The gyromagnetic ratio in electromagnetism
- Gamma waves in neuroscience
- Gamma motor neurons in neuroscience
- A non-SI metric unit of measure of mass equal to one microgram (1 μg).[7][8] This always-rare use is deprecated.
- A non-SI unit of measure of magnetic flux density, sometimes used in geophysics, equal to 10-5 Gauss (G), or 1 nanotesla (nT).[9]
- The power by which the luminance of an image is increased in gamma correction
- In civil and mechanical engineering:
- Specific weight
- The shear rate of a fluid is represented by a lowercase gamma with a dot above it:
- Austenite (also known as γ-iron), a metallic non-magnetic allotrope or solid solution of iron.
- The gamma carbon, the third carbon attached to a functional group in organic chemistry and biochemistry; see Alpha and beta carbon
The lowercase Latin gamma ɣ can also be used in contexts (such as chemical or molecule nomenclature) where gamma must not be confused with the letter y, which can occur in some computer typefaces.
Uppercase
The uppercase letter is used as a symbol for:
- In mathematics, the gamma function (usually written as -function) is an extension of the factorial to complex numbers
- In mathematics, the upper incomplete gamma function
- The Christoffel symbols in differential geometry
- In probability theory and statistics, the gamma distribution is a two-parameter family of continuous probability distributions.
- In solid-state physics, the center of the Brillouin zone
- Circulation in fluid mechanics
- As reflection coefficient in physics and electrical engineering
- The tape alphabet of a Turing machine
- The Feferman–Schütte ordinal
- One of the Greeks in mathematical finance
Meteorology
Tropical cyclones
The name Gamma has been used twice for tropical cyclones:
Tropical Storm Gamma (2005) - deadly tropical storm that impacted Honduras
Hurricane Gamma (2020) - hurricane that affected the Yucatan Peninsula
Encoding
HTML
The HTML entities for uppercase and lowercase gamma are Γ and γ.
Unicode
- Greek Gamma
| Preview | Γ | γ | ᴦ | ᵞ | ᵧ | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Unicode name | GREEK CAPITAL LETTER GAMMA | GREEK SMALL LETTER GAMMA | GREEK LETTER SMALL CAPITAL GAMMA | MODIFIER LETTER SMALL GREEK GAMMA | GREEK SUBSCRIPT SMALL LETTER GAMMA | |||||
| Encodings | decimal | hex | dec | hex | dec | hex | dec | hex | dec | hex |
| Unicode | 915 | U+0393 | 947 | U+03B3 | 7462 | U+1D26 | 7518 | U+1D5E | 7527 | U+1D67 |
| UTF-8 | 206 147 | CE 93 | 206 179 | CE B3 | 225 180 166 | E1 B4 A6 | 225 181 158 | E1 B5 9E | 225 181 167 | E1 B5 A7 |
| Numeric character reference | Γ |
Γ |
γ |
γ |
ᴦ |
ᴦ |
ᵞ |
ᵞ |
ᵧ |
ᵧ |
| Named character reference | Γ | γ |
| |||||||
- Coptic Gamma
| Preview | Ⲅ | ⲅ | ||
|---|---|---|---|---|
| Unicode name | COPTIC CAPITAL LETTER GAMMA | COPTIC SMALL LETTER GAMMA | ||
| Encodings | decimal | hex | dec | hex |
| Unicode | 11396 | U+2C84 | 11397 | U+2C85 |
| UTF-8 | 226 178 132 | E2 B2 84 | 226 178 133 | E2 B2 85 |
| Numeric character reference | Ⲅ |
Ⲅ |
ⲅ |
ⲅ |
| Preview | Ɣ | ɣ | ˠ | ɤ | ||||
|---|---|---|---|---|---|---|---|---|
| Unicode name | LATIN CAPITAL LETTER GAMMA | LATIN SMALL LETTER GAMMA | MODIFIER LETTER SMALL GAMMA | LATIN SMALL LETTER RAMS HORN | ||||
| Encodings | decimal | hex | dec | hex | dec | hex | dec | hex |
| Unicode | 404 | U+0194 | 611 | U+0263 | 736 | U+02E0 | 612 | U+0264 |
| UTF-8 | 198 148 | C6 94 | 201 163 | C9 A3 | 203 160 | CB A0 | 201 164 | C9 A4 |
| Numeric character reference | Ɣ |
Ɣ |
ɣ |
ɣ |
ˠ |
ˠ |
ɤ |
ɤ |
- CJK Square Gamma
| Preview | ㌏ | |
|---|---|---|
| Unicode name | SQUARE GAMMA | |
| Encodings | decimal | hex |
| Unicode | 13071 | U+330F |
| UTF-8 | 227 140 143 | E3 8C 8F |
| Numeric character reference | ㌏ |
㌏ |
- Technical / Mathematical Gamma
| Preview | ℾ | ℽ | 𝚪 | 𝛄 | 𝛤 | 𝛾 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unicode name | DOUBLE-STRUCK CAPITAL GAMMA |
DOUBLE-STRUCK SMALL GAMMA |
MATHEMATICAL BOLD CAPITAL GAMMA |
MATHEMATICAL BOLD SMALL GAMMA |
MATHEMATICAL ITALIC CAPITAL GAMMA |
MATHEMATICAL ITALIC SMALL GAMMA | ||||||
| Encodings | decimal | hex | dec | hex | dec | hex | dec | hex | dec | hex | dec | hex |
| Unicode | 8510 | U+213E | 8509 | U+213D | 120490 | U+1D6AA | 120516 | U+1D6C4 | 120548 | U+1D6E4 | 120574 | U+1D6FE |
| UTF-8 | 226 132 190 | E2 84 BE | 226 132 189 | E2 84 BD | 240 157 154 170 | F0 9D 9A AA | 240 157 155 132 | F0 9D 9B 84 | 240 157 155 164 | F0 9D 9B A4 | 240 157 155 190 | F0 9D 9B BE |
| UTF-16 | 8510 | 213E | 8509 | 213D | 55349 57002 | D835 DEAA | 55349 57028 | D835 DEC4 | 55349 57060 | D835 DEE4 | 55349 57086 | D835 DEFE |
| Numeric character reference | ℾ |
ℾ |
ℽ |
ℽ |
𝚪 |
𝚪 |
𝛄 |
𝛄 |
𝛤 |
𝛤 |
𝛾 |
𝛾 |
| Preview | 𝜞 | 𝜸 | 𝝘 | 𝝲 | 𝞒 | 𝞬 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Unicode name | MATHEMATICAL BOLD ITALIC CAPITAL GAMMA |
MATHEMATICAL BOLD ITALIC SMALL GAMMA |
MATHEMATICAL SANS-SERIF BOLD CAPITAL GAMMA |
MATHEMATICAL SANS-SERIF BOLD SMALL GAMMA |
MATHEMATICAL SANS-SERIF BOLD ITALIC CAPITAL GAMMA |
MATHEMATICAL SANS-SERIF BOLD ITALIC SMALL GAMMA | ||||||
| Encodings | decimal | hex | dec | hex | dec | hex | dec | hex | dec | hex | dec | hex |
| Unicode | 120606 | U+1D71E | 120632 | U+1D738 | 120664 | U+1D758 | 120690 | U+1D772 | 120722 | U+1D792 | 120748 | U+1D7AC |
| UTF-8 | 240 157 156 158 | F0 9D 9C 9E | 240 157 156 184 | F0 9D 9C B8 | 240 157 157 152 | F0 9D 9D 98 | 240 157 157 178 | F0 9D 9D B2 | 240 157 158 146 | F0 9D 9E 92 | 240 157 158 172 | F0 9D 9E AC |
| UTF-16 | 55349 57118 | D835 DF1E | 55349 57144 | D835 DF38 | 55349 57176 | D835 DF58 | 55349 57202 | D835 DF72 | 55349 57234 | D835 DF92 | 55349 57260 | D835 DFAC |
| Numeric character reference | 𝜞 |
𝜞 |
𝜸 |
𝜸 |
𝝘 |
𝝘 |
𝝲 |
𝝲 |
𝞒 |
𝞒 |
𝞬 |
𝞬 |
These characters are used only as mathematical symbols. Stylized Greek text should be encoded using the normal Greek letters, with markup and formatting to indicate text style.
See also
References
that fateful 100 gamma, the same dosage I had had at my first LSD session
- Weisstein, Eric W. (30 April 2023). "Gamma -- from Eric Weisstein's World of Physics". scienceworld.wolfram.com.
https://en.wikipedia.org/wiki/Gamma
| Hairy cell leukemia | |
|---|---|
 | |
| Specialty | Hematology and oncology |
Hairy cell leukemia is an uncommon hematological malignancy characterized by an accumulation of abnormal B lymphocytes.[1] It is usually classified as a subtype of chronic lymphocytic leukemia (CLL). Hairy cell leukemia makes up about 2% of all leukemias, with fewer than 2,000 new cases diagnosed annually in North America and Western Europe combined.
Hairy cell leukemia (HCL) was originally described as histiocytic leukemia, malignant reticulosis, or lymphoid myelofibrosis in publications dating back to the 1920s. The disease was formally named leukemic reticuloendotheliosis, and its characterization was significantly advanced by Bertha Bouroncle and colleagues at the Ohio State University College of Medicine in 1958. Its common name, which was coined in 1966,[2] is derived from the "hairy" appearance of the malignant B cells under a microscope.[3]
https://en.wikipedia.org/wiki/Hairy_cell_leukemia
Hair cells are the sensory receptors of both the auditory system and the vestibular system in the ears of all vertebrates, and in the lateral line organ of fishes. Through mechanotransduction, hair cells detect movement in their environment.[1]
In mammals, the auditory hair cells are located within the spiral organ of Corti on the thin basilar membrane in the cochlea of the inner ear. They derive their name from the tufts of stereocilia called hair bundles that protrude from the apical surface of the cell into the fluid-filled cochlear duct. The stereocilia number from fifty to a hundred in each cell while being tightly packed together[2] and decrease in size the further away they are located from the kinocilium.[3] The hair bundles are arranged as stiff columns that move at their base in response to stimuli applied to the tips.[4]
Mammalian cochlear hair cells are of two anatomically and functionally distinct types, known as outer, and inner hair cells. Damage to these hair cells results in decreased hearing sensitivity, and because the inner ear hair cells cannot regenerate, this damage is permanent.[5] Damage to hair cells can cause damage to the vestibular system and therefore causing difficulties in balancing. However, other organisms, such as the frequently studied zebrafish, and birds have hair cells that can regenerate.[6][7] The human cochlea contains on the order of 3,500 inner hair cells and 12,000 outer hair cells at birth.[8]
The outer hair cells mechanically amplify low-level sound that enters the cochlea.[9][10] The amplification may be powered by the movement of their hair bundles, or by an electrically driven motility of their cell bodies. This so-called somatic electromotility amplifies sound in all land vertebrates. It is affected by the closing mechanism of the mechanical sensory ion channels at the tips of the hair bundles.[citation needed]
The inner hair cells transform the sound vibrations in the fluids of the cochlea into electrical signals that are then relayed via the auditory nerve to the auditory brainstem and to the auditory cortex.
https://en.wikipedia.org/wiki/Hair_cell
Inner hair cells – from sound to nerve signal
The deflection of the hair-cell stereocilia opens mechanically gated ion channels that allow any small, positively charged ions (primarily potassium and calcium) to enter the cell.[11] Unlike many other electrically active cells, the hair cell itself does not fire an action potential. Instead, the influx of positive ions from the endolymph in the scala media depolarizes the cell, resulting in a receptor potential. This receptor potential opens voltage gated calcium channels; calcium ions then enter the cell and trigger the release of neurotransmitters at the basal end of the cell. The neurotransmitters diffuse across the narrow space between the hair cell and a nerve terminal, where they then bind to receptors and thus trigger action potentials in the nerve. In this way, the mechanical sound signal is converted into an electrical nerve signal. Repolarization of hair cells is done in a special manner. The perilymph in the scala tympani has a very low concentration of positive ions. The electrochemical gradient makes the positive ions flow through channels to the perilymph.
Hair cells chronically leak Ca2+. This leakage causes a tonic release of neurotransmitter to the synapses. It is thought that this tonic release is what allows the hair cells to respond so quickly in response to mechanical stimuli. The quickness of the hair cell response may also be due to the fact that it can increase the amount of neurotransmitter release in response to a change of as little as 100 μV in membrane potential.[12]
Hair cells are also able to distinguish tone frequencies through one of two methods. The first method, found only in non-mammals, uses electrical resonance in the basolateral membrane of the hair cell. The electrical resonance for this method appears as a damped oscillation of membrane potential responding to an applied current pulse. The second method uses tonotopic differences of the basilar membrane. This difference comes from the different locations of the hair cells. Hair cells that have high-frequency resonance are located at the basal end while hair cells that have significantly lower frequency resonance are found at the apical end of the epithelium.[13]
Outer hair cells – acoustical pre-amplifiers
In mammalian outer hair cells, the varying receptor potential is converted to active vibrations of the cell body. This mechanical response to electrical signals is termed somatic electromotility;[14] it drives variations in the cell's length, synchronized to the incoming sound signal, and provides mechanical amplification by feedback to the traveling wave.[15]
Outer hair cells are found only in mammals. While hearing sensitivity of mammals is similar to that of other classes of vertebrates, without functioning outer hair cells, the sensitivity decreases by approximately 50 dB.[16] Outer hair cells extend the hearing range to about 200 kHz in some marine mammals.[17] They have also improved frequency selectivity (frequency discrimination), which is of particular benefit for humans, because it enabled sophisticated speech and music. Outer hair cells are functional even after cellular stores of ATP are depleted.[14]
The effect of this system is to nonlinearly amplify quiet sounds more than large ones so that a wide range of sound pressures can be reduced to a much smaller range of hair displacements.[18] This property of amplification is called the cochlear amplifier.
The molecular biology of hair cells has seen considerable progress in recent years, with the identification of the motor protein (prestin) that underlies somatic electromotility in the outer hair cells. Prestin's function has been shown to be dependent on chloride channel signaling and that it is compromised by the common marine pesticide tributyltin. Because this class of pollutant bioconcentrates up the food chain, the effect is pronounced in top marine predators such as orcas and toothed whales.[19]
Hair cell signal adaption
Calcium ion influx plays an important role for the hair cells to adapt to the amplification of the signal. This allows humans to ignore constant sounds that are no longer new and allow us to be acute to other changes in our surrounding. The key adaptation mechanism comes from a motor protein myosin-1c that allows slow adaptation, provides tension to sensitize transduction channels, and also participate in signal transduction apparatus.[20][21] More recent research now shows that the calcium-sensitive binding of calmodulin to myosin-1c could actually modulate the interaction of the adaptation motor with other components of the transduction apparatus as well.[22][23]
Fast Adaptation: During fast adaptation, Ca2+ ions that enter a stereocilium through an open MET channel bind rapidly to a site on or near the channel and induce channel closure. When channels close, tension increases in the tip link, pulling the bundle in the opposite direction.[20] Fast adaptation is more prominent in sound and auditory detecting hair cells, rather in vestibular cells.
Slow Adaption: The dominating model suggests that slow adaptation occurs when myosin-1c slides down the stereocilium in response to elevated tension during bundle displacement.[20] The resultant decreased tension in the tip link permits the bundle to move farther in the opposite direction. As tension decreases, channels close, producing the decline in transduction current.[20] Slow adaptation is most prominent in vestibular hair cells that sense spatial movement and less in cochlear hair cells that detect auditory signals.[21]
Neural connection
This section needs additional citations for verification. (September 2016) |
Neurons of the auditory or vestibulocochlear nerve (the eighth cranial nerve) innervate cochlear and vestibular hair cells.[24] The neurotransmitter released by hair cells that stimulates the terminal neurites of peripheral axons of the afferent (towards the brain) neurons is thought to be glutamate. At the presynaptic juncture, there is a distinct presynaptic dense body or ribbon. This dense body is surrounded by synaptic vesicles and is thought to aid in the fast release of neurotransmitter.
Nerve fiber innervation is much denser for inner hair cells than for outer hair cells. A single inner hair cell is innervated by numerous nerve fibers, whereas a single nerve fiber innervates many outer hair cells. Inner hair cell nerve fibers are also very heavily myelinated, which is in contrast to the unmyelinated outer hair cell nerve fibers. The region of the basilar membrane supplying the inputs to a particular afferent nerve fibre can be considered to be its receptive field.
Efferent projections from the brain to the cochlea also play a role in the perception of sound. Efferent synapses occur on outer hair cells and on afferent axons under inner hair cells. The presynaptic terminal bouton is filled with vesicles containing acetylcholine and a neuropeptide called calcitonin gene-related peptide. The effects of these compounds vary; in some hair cells the acetylcholine hyperpolarizes the cell, which reduces the sensitivity of the cochlea locally.
Regrowth
Research on the regrowth of cochlear cells may lead to medical treatments that restore hearing. Unlike birds and fish, humans and other mammals are generally incapable of regrowing the cells of the inner ear that convert sound into neural signals when those cells are damaged by age or disease.[7][25] Researchers are making progress in gene therapy and stem-cell therapy that may allow the damaged cells to be regenerated. Because hair cells of auditory and vestibular systems in birds and fish have been found to regenerate, their ability has been studied at length.[7][26] In addition, lateral line hair cells, which have a mechanotransduction function, have been shown to regrow in organisms, such as the zebrafish.[27]
Researchers have identified a mammalian gene that normally acts as a molecular switch to block the regrowth of cochlear hair cells in adults.[28] The Rb1 gene encodes the retinoblastoma protein, which is a tumor suppressor. Rb stops cells from dividing by encouraging their exit from the cell cycle.[29][30] Not only do hair cells in a culture dish regenerate when the Rb1 gene is deleted, but mice bred to be missing the gene grow more hair cells than control mice that have the gene. Additionally, the sonic hedgehog protein has been shown to block activity of the retinoblastoma protein, thereby inducing cell cycle re-entry and the regrowth of new cells.[31]
Several Notch signaling pathway inhibitors, including the gamma secretase inhibitor LY3056480, are being studied for their potential ability to regenerate hair cells in the cochlea.[32][33]
TBX2 (T-box transcription factor 2) has been shown to be a master regulator in the differentiation of inner and outer hair cells.[34] This discovery has allowed researchers to direct hair cells to develop into either inner or outer hair cells, which could help in replacing hair cells that have died and prevent or reverse hearing loss.[35][36]
The cell cycle inhibitor p27kip1 (CDKN1B) has also been found to encourage regrowth of cochlear hair cells in mice following genetic deletion or knock down with siRNA targeting p27.[37][38] Research on hair cell regeneration may bring us closer to clinical treatment for human hearing loss caused by hair cell damage or death.
Additional images
The lamina reticularis and subjacent structures.
References
- Ono K, Nakagawa T, Kojima K, Matsumoto M, Kawauchi T, Hoshino M, Ito J (Dec 2009). "Silencing p27 reverses post-mitotic state of supporting cells in neonatal mouse cochleae" (PDF). Mol Cell Neurosci. 42 (4): 391–8. doi:10.1016/j.mcn.2009.08.011. hdl:2433/87734. PMID 19733668. S2CID 206831997. (primary source)
Bibliography
- Coffin A, Kelley M, Manley GA, Popper AN (2004). "Evolution of sensory hair cells". In Manley, et al. (eds.). Evolution of the Vertebrate Auditory System. pp. 55–94.
- Fettiplace R, Hackney CM (2006). "The sensory and motor roles of auditory hair cells". Nature Reviews. Neuroscience. 7 (1): 19–29. doi:10.1038/nrn1828. PMID 16371947. S2CID 10155096.
- Kandel ER, Schwartz JH, Jessell TM (2000). Principles of Neural Science (4th ed.). New York: McGraw-Hill. pp. 590–594. ISBN 0-8385-7701-6.
- Manley GA, Popper AN, Fay RR (2004). Evolution of the Vertebrate Auditory System. New York: Springer-Verlag. ISBN 0-387-21093-8.
- Manley GA (2004). "Advances and perspectives in the study of the evolution of the vertebrate auditory system". In Manley, et al. (eds.). Evolution of the Vertebrate Auditory System. pp. 360–368.
- Rabbitt RD, Boyle R, Highstein SM (1–5 February 2010). "Mechanical amplification by hair cells in the semicircular canals". Proceedings of the National Academy of Sciences. 107 (8): 3864–9. Bibcode:2010PNAS..107.3864R. doi:10.1073/pnas.0906765107. PMC 2840494. PMID 20133682.
- "Built-in amps: How subtle head motions, quiet sounds are reported to the brain". Medical Xpress. February 9, 2010.
- Breneman KD, Brownell WE, Rabbitt RD (22 April 2009). Brezina V (ed.). "Hair cell bundles: flexoelectric motors of the inner ear". PLOS ONE. 4 (4): e5201. Bibcode:2009PLoSO...4.5201B. doi:10.1371/journal.pone.0005201. PMC 2668172. PMID 19384413.
- "Power steering for your hearing: Ears have tiny 'flexoelectric' motors to amplify sound". Phys.org (Press release). April 22, 2009.
External links
- Molecular Basis of Hearing
- Outer hair cell dancing "rock around the clock"
- Dancing OHC video Yale Ear Lab
- NIF Search – Hair Cell via the Neuroscience Information Framework
- Hair-Tuning-Sound-Sensor Archived 2021-08-26 at the Wayback Machine A concise report on the recent development of sound sensors based on hair tuning by students of SMMEE, IIT Ropar
- Auditory system
- Receptor cells
- Human cells
https://en.wikipedia.org/wiki/Hair_cell
The hairy yellow-shouldered bat (Sturnira erythromos) is a species of bat in the family Phyllostomidae native to South America. There are no recognised subspecies. - https://en.wikipedia.org/wiki/Hairy_yellow-shouldered_bat
The patagium (plural: patagia) is a membranous body part that assists an animal in obtaining lift when gliding or flight. The structure is found in extant and extinct groups of flying and gliding animals including bats, birds, some dromaeosaurs, pterosaurs, gliding mammals, some flying lizards, and flying frogs. The patagium that stretches between an animal's hind limbs is called the uropatagium (especially in bats) or the interfemoral membrane.
https://en.wikipedia.org/wiki/Patagium
Draco is a genus of agamid lizards[1] that are also known as flying lizards, flying dragons or gliding lizards. These lizards are capable of gliding flight via membranes that may be extended to create wings (patagia), formed by an enlarged set of ribs. They are arboreal insectivores.
While not capable of powered flight they often obtain lift in the course of their gliding flights. Glides as long as 60 m (200 ft) have been recorded, over which the animal loses only 10 m (33 ft) in height, which is quite some distance, considering that one lizard is only around 20 cm (7.9 in) in total length, tail included.[2] They are found across Southeast Asia and southern India and are fairly common in forests, areca gardens, teak plantations and shrub jungle.
| Draco | |
|---|---|

| |
| D. taeniopterus in mid-glide, on Bulon Island, Thailand |
https://en.wikipedia.org/wiki/Draco_(lizard)
An insectivore is a carnivorous animal or plant that eats insects.[1] An alternative term is entomophage,[2] which can also refer to the human practice of eating insects.
https://en.wikipedia.org/wiki/Insectivore
V600E is a mutation of the BRAF gene in which valine (V) is substituted by glutamic acid (E) at amino acid 600.[1][2] It is a driver mutation in a proportion of certain diagnoses, including melanoma,[3][4] hairy cell leukemia,[5][6] papillary thyroid carcinoma,[7][8] colorectal cancer,[9] non-small-cell lung cancer,[10][11] Langerhans cell histiocytosis,[12] Erdheim–Chester disease (a non-Langerhans-cell histiocytosis) and ameloblastoma.[13]
The mechanism of the mutation is that the negative charge of the acidic glutamic acid residue causes it to be phosphomimetic. This mimics the phosphorylation of the nearby T599 threonine and S602 serine residues in the activation segment of BRAF, which are used to activate the wild type form of the protein. The glutamate residue of the mutant therefore functions to activate BRAF by inhibiting the interaction of the BRAF's glycine rich loop and activation segment, which would ordinarily be inhibitory. The loss of inhibition of BRAF leads to an increase in its basal activity and hence is oncogenic.
Clinical
Vemurafenib, encorafenib, and dabrafenib are approved by the FDA for treatment of metastatic melanomas that express V600E.
https://en.wikipedia.org/wiki/V600E
Phosphomimetics are amino acid substitutions that mimic a phosphorylated protein, thereby activating (or deactivating) the protein. Within cells, proteins are commonly modified at serine, tyrosine and threonine amino acids by adding a phosphate group. Phosphorylation is a common mode of activating or deactivating a protein as a form of regulation. However some non-phosphorylated amino acids appear chemically similar to phosphorylated amino acids. Therefore, by replacing an amino acid, the protein may maintain a higher level of activity. For example, aspartic acid is chemically similar to phospho-serine. Therefore, when an aspartic acid replaces a serine, it is a phosphomimetic of phospho-serine and can make the protein always in its phosphorylated form. Phosphonate-based compounds have been used as phosphotyrosine analogues, as they are less enzyme labile and are physiologically more stable.[1]
https://en.wikipedia.org/wiki/Phosphomimetics
By effect on function
A mutation becomes an effect on function mutation when the exactitude of functions between a mutated protein and its direct interactor undergoes change. The interactors can be other proteins, molecules, nucleic acids, etc. There are many mutations that fall under the category of by effect on function, but depending on the specificity of the change the mutations listed below will occur.[50]
- Loss-of-function mutations, also called inactivating mutations, result in the gene product having less or no function (being partially or wholly inactivated). When the allele has a complete loss of function (null allele), it is often called an amorph or amorphic mutation in Muller's morphs schema. Phenotypes associated with such mutations are most often recessive. Exceptions are when the organism is haploid, or when the reduced dosage of a normal gene product is not enough for a normal phenotype (this is called haploinsufficiency). A disease that is caused by a loss-of-function mutation is Gitelman syndrome and cystic fibrosis.[51]
- Gain-of-function mutations also called activating mutations, change the gene product such that its effect gets stronger (enhanced activation) or even is superseded by a different and abnormal function. When the new allele is created, a heterozygote containing the newly created allele as well as the original will express the new allele; genetically this defines the mutations as dominant phenotypes. Several of Muller's morphs correspond to the gain of function, including hypermorph (increased gene expression) and neomorph (novel function).
- Dominant negative mutations (also called anti-morphic mutations) have an altered gene product that acts antagonistically to the wild-type allele. These mutations usually result in an altered molecular function (often inactive) and are characterized by a dominant or semi-dominant phenotype. In humans, dominant negative mutations have been implicated in cancer (e.g., mutations in genes p53, ATM, CEBPA, and PPARgamma). Marfan syndrome is caused by mutations in the FBN1 gene, located on chromosome 15, which encodes fibrillin-1, a glycoprotein component of the extracellular matrix. Marfan syndrome is also an example of dominant negative mutation and haploinsufficiency.
- Lethal mutations result in the instant death of the developing organism. Lethal mutations can also lead to a substantial loss in the life expectancy of the organism. An example of a disease that is caused by a dominant lethal mutation is Huntington's disease.
- Null mutations, also known as Amorphic mutations, are a form of loss-of-function mutations that completely prohibit the gene's function. The mutation leads to a complete loss of operation at the phenotypic level, also causing no gene product to be formed. Atopic eczema and dermatitis syndrome are common diseases caused by a null mutation of the gene that activates filaggrin.
- Suppressor mutations are a type of mutation that causes the double mutation to appear normally. In suppressor mutations the phenotypic activity of a different mutation is completely suppressed, thus causing the double mutation to look normal. There are two types of suppressor mutations, there are intragenic and extragenic suppressor mutations. Intragenic mutations occur in the gene where the first mutation occurs, while extragenic mutations occur in the gene that interacts with the product of the first mutation. A common disease that results from this type of mutation is Alzheimer's disease.[52]
- Neomorphic mutations are a part of the gain-of-function mutations and are characterized by the control of new protein product synthesis. The newly synthesized gene normally contains a novel gene expression or molecular function. The result of the neomorphic mutation is the gene where the mutation occurs has a complete change in function.[53]
- A back mutation or reversion is a point mutation that restores the original sequence and hence the original phenotype.[54]
By effect on fitness (harmful, beneficial, neutral mutations)
In genetics, it is sometimes useful to classify mutations as either harmful or beneficial (or neutral):
- A harmful, or deleterious, mutation decreases the fitness of the organism. Many, but not all mutations in essential genes are harmful (if a mutation does not change the amino acid sequence in an essential protein, it is harmless in most cases).
- A beneficial, or advantageous mutation increases the fitness of the organism. Examples are mutations that lead to antibiotic resistance in bacteria (which are beneficial for bacteria but usually not for humans).
- A neutral mutation has no harmful or beneficial effect on the organism. Such mutations occur at a steady rate, forming the basis for the molecular clock. In the neutral theory of molecular evolution, neutral mutations provide genetic drift as the basis for most variation at the molecular level. In animals or plants, most mutations are neutral, given that the vast majority of their genomes is either non-coding or consists of repetitive sequences that have no obvious function ("junk DNA").[55]
Large-scale quantitative mutagenesis screens, in which thousands of millions of mutations are tested, invariably find that a larger fraction of mutations has harmful effects but always returns a number of beneficial mutations as well. For instance, in a screen of all gene deletions in E. coli, 80% of mutations were negative, but 20% were positive, even though many had a very small effect on growth (depending on condition).[56] Note that gene deletions involve removal of whole genes, so that point mutations almost always have a much smaller effect. In a similar screen in Streptococcus pneumoniae, but this time with transposon insertions, 76% of insertion mutants were classified as neutral, 16% had a significantly reduced fitness, but 6% were advantageous.[57]
This classification is obviously relative and somewhat artificial: a harmful mutation can quickly turn into a beneficial mutations when conditions change. Also, there is a gradient from harmful/beneficial to neutral, as many mutations may have small and mostly neglectable effects but under certain conditions will become relevant. Also, many traits are determined by hundreds of genes (or loci), so that each locus has only a minor effect. For instance, human height is determined by hundreds of genetic variants ("mutations") but each of them has a very minor effect on height,[58] apart from the impact of nutrition. Height (or size) itself may be more or less beneficial as the huge range of sizes in animal or plant groups shows.
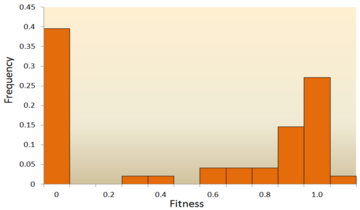
Distribution of fitness effects (DFE)
Attempts have been made to infer the distribution of fitness effects (DFE) using mutagenesis experiments and theoretical models applied to molecular sequence data. DFE, as used to determine the relative abundance of different types of mutations (i.e., strongly deleterious, nearly neutral or advantageous), is relevant to many evolutionary questions, such as the maintenance of genetic variation,[59] the rate of genomic decay,[60] the maintenance of outcrossing sexual reproduction as opposed to inbreeding[61] and the evolution of sex and genetic recombination.[62] DFE can also be tracked by tracking the skewness of the distribution of mutations with putatively severe effects as compared to the distribution of mutations with putatively mild or absent effect.[63] In summary, the DFE plays an important role in predicting evolutionary dynamics.[64][65] A variety of approaches have been used to study the DFE, including theoretical, experimental and analytical methods.
- Mutagenesis experiment: The direct method to investigate the DFE is to induce mutations and then measure the mutational fitness effects, which has already been done in viruses, bacteria, yeast, and Drosophila. For example, most studies of the DFE in viruses used site-directed mutagenesis to create point mutations and measure relative fitness of each mutant.[66][67][68][69] In Escherichia coli, one study used transposon mutagenesis to directly measure the fitness of a random insertion of a derivative of Tn10.[70] In yeast, a combined mutagenesis and deep sequencing approach has been developed to generate high-quality systematic mutant libraries and measure fitness in high throughput.[71] However, given that many mutations have effects too small to be detected[72] and that mutagenesis experiments can detect only mutations of moderately large effect; DNA sequence analysis can provide valuable information about these mutations.
- Molecular sequence analysis: With rapid development of DNA sequencing technology, an enormous amount of DNA sequence data is available and even more is forthcoming in the future. Various methods have been developed to infer the DFE from DNA sequence data.[73][74][75][76] By examining DNA sequence differences within and between species, we are able to infer various characteristics of the DFE for neutral, deleterious and advantageous mutations.[24] To be specific, the DNA sequence analysis approach allows us to estimate the effects of mutations with very small effects, which are hardly detectable through mutagenesis experiments.This figure shows a simplified version of loss-of-function, switch-of-function, gain-of-function, and conservation-of-function mutations.
One of the earliest theoretical studies of the distribution of fitness effects was done by Motoo Kimura, an influential theoretical population geneticist. His neutral theory of molecular evolution proposes that most novel mutations will be highly deleterious, with a small fraction being neutral.[25][77] A later proposal by Hiroshi Akashi proposed a bimodal model for the DFE, with modes centered around highly deleterious and neutral mutations.[78] Both theories agree that the vast majority of novel mutations are neutral or deleterious and that advantageous mutations are rare, which has been supported by experimental results. One example is a study done on the DFE of random mutations in vesicular stomatitis virus.[66] Out of all mutations, 39.6% were lethal, 31.2% were non-lethal deleterious, and 27.1% were neutral. Another example comes from a high throughput mutagenesis experiment with yeast.[71] In this experiment it was shown that the overall DFE is bimodal, with a cluster of neutral mutations, and a broad distribution of deleterious mutations.
Though relatively few mutations are advantageous, those that are play an important role in evolutionary changes.[79] Like neutral mutations, weakly selected advantageous mutations can be lost due to random genetic drift, but strongly selected advantageous mutations are more likely to be fixed. Knowing the DFE of advantageous mutations may lead to increased ability to predict the evolutionary dynamics. Theoretical work on the DFE for advantageous mutations has been done by John H. Gillespie[80] and H. Allen Orr.[81] They proposed that the distribution for advantageous mutations should be exponential under a wide range of conditions, which, in general, has been supported by experimental studies, at least for strongly selected advantageous mutations.[82][83][84]
In general, it is accepted that the majority of mutations are neutral or deleterious, with advantageous mutations being rare; however, the proportion of types of mutations varies between species. This indicates two important points: first, the proportion of effectively neutral mutations is likely to vary between species, resulting from dependence on effective population size; second, the average effect of deleterious mutations varies dramatically between species.[24] In addition, the DFE also differs between coding regions and noncoding regions, with the DFE of noncoding DNA containing more weakly selected mutations.[24]
By inheritance
In multicellular organisms with dedicated reproductive cells, mutations can be subdivided into germline mutations, which can be passed on to descendants through their reproductive cells, and somatic mutations (also called acquired mutations),[85] which involve cells outside the dedicated reproductive group and which are not usually transmitted to descendants.
Diploid organisms (e.g., humans) contain two copies of each gene—a paternal and a maternal allele. Based on the occurrence of mutation on each chromosome, we may classify mutations into three types. A wild type or homozygous non-mutated organism is one in which neither allele is mutated.
- A heterozygous mutation is a mutation of only one allele.
- A homozygous mutation is an identical mutation of both the paternal and maternal alleles.
- Compound heterozygous mutations or a genetic compound consists of two different mutations in the paternal and maternal alleles.[86]
Germline mutation
A germline mutation in the reproductive cells of an individual gives rise to a constitutional mutation in the offspring, that is, a mutation that is present in every cell. A constitutional mutation can also occur very soon after fertilisation, or continue from a previous constitutional mutation in a parent.[87] A germline mutation can be passed down through subsequent generations of organisms.
The distinction between germline and somatic mutations is important in animals that have a dedicated germline to produce reproductive cells. However, it is of little value in understanding the effects of mutations in plants, which lack a dedicated germline. The distinction is also blurred in those animals that reproduce asexually through mechanisms such as budding, because the cells that give rise to the daughter organisms also give rise to that organism's germline.
A new germline mutation not inherited from either parent is called a de novo mutation.
Somatic mutation
A change in the genetic structure that is not inherited from a parent, and also not passed to offspring, is called a somatic mutation.[85] Somatic mutations are not inherited by an organism's offspring because they do not affect the germline. However, they are passed down to all the progeny of a mutated cell within the same organism during mitosis. A major section of an organism therefore might carry the same mutation. These types of mutations are usually prompted by environmental causes, such as ultraviolet radiation or any exposure to certain harmful chemicals, and can cause diseases including cancer.[88]
With plants, some somatic mutations can be propagated without the need for seed production, for example, by grafting and stem cuttings. These type of mutation have led to new types of fruits, such as the "Delicious" apple and the "Washington" navel orange.[89]
Human and mouse somatic cells have a mutation rate more than ten times higher than the germline mutation rate for both species; mice have a higher rate of both somatic and germline mutations per cell division than humans. The disparity in mutation rate between the germline and somatic tissues likely reflects the greater importance of genome maintenance in the germline than in the soma.[90]
Special classes
- Conditional mutation is a mutation that has wild-type (or less severe) phenotype under certain "permissive" environmental conditions and a mutant phenotype under certain "restrictive" conditions. For example, a temperature-sensitive mutation can cause cell death at high temperature (restrictive condition), but might have no deleterious consequences at a lower temperature (permissive condition).[91] These mutations are non-autonomous, as their manifestation depends upon presence of certain conditions, as opposed to other mutations which appear autonomously.[92] The permissive conditions may be temperature,[93] certain chemicals,[94] light[94] or mutations in other parts of the genome.[92] In vivo mechanisms like transcriptional switches can create conditional mutations. For instance, association of Steroid Binding Domain can create a transcriptional switch that can change the expression of a gene based on the presence of a steroid ligand.[95] Conditional mutations have applications in research as they allow control over gene expression. This is especially useful studying diseases in adults by allowing expression after a certain period of growth, thus eliminating the deleterious effect of gene expression seen during stages of development in model organisms.[94] DNA Recombinase systems like Cre-Lox recombination used in association with promoters that are activated under certain conditions can generate conditional mutations. Dual Recombinase technology can be used to induce multiple conditional mutations to study the diseases which manifest as a result of simultaneous mutations in multiple genes.[94] Certain inteins have been identified which splice only at certain permissive temperatures, leading to improper protein synthesis and thus, loss-of-function mutations at other temperatures.[96] Conditional mutations may also be used in genetic studies associated with ageing, as the expression can be changed after a certain time period in the organism's lifespan.[93]
- Replication timing quantitative trait loci affects DNA replication.
Nomenclature
In order to categorize a mutation as such, the "normal" sequence must be obtained from the DNA of a "normal" or "healthy" organism (as opposed to a "mutant" or "sick" one), it should be identified and reported; ideally, it should be made publicly available for a straightforward nucleotide-by-nucleotide comparison, and agreed upon by the scientific community or by a group of expert geneticists and biologists, who have the responsibility of establishing the standard or so-called "consensus" sequence. This step requires a tremendous scientific effort. Once the consensus sequence is known, the mutations in a genome can be pinpointed, described, and classified. The committee of the Human Genome Variation Society (HGVS) has developed the standard human sequence variant nomenclature,[97] which should be used by researchers and DNA diagnostic centers to generate unambiguous mutation descriptions. In principle, this nomenclature can also be used to describe mutations in other organisms. The nomenclature specifies the type of mutation and base or amino acid changes.
- Nucleotide substitution (e.g., 76A>T) – The number is the
position of the nucleotide from the 5' end; the first letter represents
the wild-type nucleotide, and the second letter represents the
nucleotide that replaced the wild type. In the given example, the
adenine at the 76th position was replaced by a thymine.
- If it becomes necessary to differentiate between mutations in genomic DNA, mitochondrial DNA, and RNA, a simple convention is used. For example, if the 100th base of a nucleotide sequence mutated from G to C, then it would be written as g.100G>C if the mutation occurred in genomic DNA, m.100G>C if the mutation occurred in mitochondrial DNA, or r.100g>c if the mutation occurred in RNA. Note that, for mutations in RNA, the nucleotide code is written in lower case.
- Amino acid substitution (e.g., D111E) – The first letter is the one letter code of the wild-type amino acid, the number is the position of the amino acid from the N-terminus, and the second letter is the one letter code of the amino acid present in the mutation. Nonsense mutations are represented with an X for the second amino acid (e.g. D111X).
- Amino acid deletion (e.g., ΔF508) – The Greek letter Δ (delta) indicates a deletion. The letter refers to the amino acid present in the wild type and the number is the position from the N terminus of the amino acid were it to be present as in the wild type.
Mutation rates
Mutation rates vary substantially across species, and the evolutionary forces that generally determine mutation are the subject of ongoing investigation.
In humans, the mutation rate is about 50-90 de novo mutations per genome per generation, that is, each human accumulates about 50-90 novel mutations that were not present in his or her parents. This number has been established by sequencing thousands of human trios, that is, two parents and at least one child.[98]
The genomes of RNA viruses are based on RNA rather than DNA. The RNA viral genome can be double-stranded (as in DNA) or single-stranded. In some of these viruses (such as the single-stranded human immunodeficiency virus), replication occurs quickly, and there are no mechanisms to check the genome for accuracy. This error-prone process often results in mutations.
Randomness of mutations
There is a widespread assumption that mutations are (entirely) "random" with respect to their consequences (in terms of probability). This was shown to be wrong as mutation frequency can vary across regions of the genome, with such DNA repair- and mutation-biases being associated with various factors. For instance, biologically important regions were found to be protected from mutations and mutations beneficial to the studied plant were found to be more likely – i.e. mutation is "non-random in a way that benefits the plant".[99][100]
Disease causation
Changes in DNA caused by mutation in a coding region of DNA can cause errors in protein sequence that may result in partially or completely non-functional proteins. Each cell, in order to function correctly, depends on thousands of proteins to function in the right places at the right times. When a mutation alters a protein that plays a critical role in the body, a medical condition can result. One study on the comparison of genes between different species of Drosophila suggests that if a mutation does change a protein, the mutation will most likely be harmful, with an estimated 70 percent of amino acid polymorphisms having damaging effects, and the remainder being either neutral or weakly beneficial.[8] Some mutations alter a gene's DNA base sequence but do not change the protein made by the gene. Studies have shown that only 7% of point mutations in noncoding DNA of yeast are deleterious and 12% in coding DNA are deleterious. The rest of the mutations are either neutral or slightly beneficial.[101]
Inherited disorders
If a mutation is present in a germ cell, it can give rise to offspring that carries the mutation in all of its cells. This is the case in hereditary diseases. In particular, if there is a mutation in a DNA repair gene within a germ cell, humans carrying such germline mutations may have an increased risk of cancer. A list of 34 such germline mutations is given in the article DNA repair-deficiency disorder. An example of one is albinism, a mutation that occurs in the OCA1 or OCA2 gene. Individuals with this disorder are more prone to many types of cancers, other disorders and have impaired vision.
DNA damage can cause an error when the DNA is replicated, and this error of replication can cause a gene mutation that, in turn, could cause a genetic disorder. DNA damages are repaired by the DNA repair system of the cell. Each cell has a number of pathways through which enzymes recognize and repair damages in DNA. Because DNA can be damaged in many ways, the process of DNA repair is an important way in which the body protects itself from disease. Once DNA damage has given rise to a mutation, the mutation cannot be repaired.
Role in carcinogenesis
On the other hand, a mutation may occur in a somatic cell of an organism. Such mutations will be present in all descendants of this cell within the same organism. The accumulation of certain mutations over generations of somatic cells is part of cause of malignant transformation, from normal cell to cancer cell.[102]
Cells with heterozygous loss-of-function mutations (one good copy of gene and one mutated copy) may function normally with the unmutated copy until the good copy has been spontaneously somatically mutated. This kind of mutation happens often in living organisms, but it is difficult to measure the rate. Measuring this rate is important in predicting the rate at which people may develop cancer.[103]
Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of mutation may be increased by mutagens. Mutagens can be physical, such as radiation from UV rays, X-rays or extreme heat, or chemical (molecules that misplace base pairs or disrupt the helical shape of DNA). Mutagens associated with cancers are often studied to learn about cancer and its prevention.
Prion mutations
Prions are proteins and do not contain genetic material. However, prion replication has been shown to be subject to mutation and natural selection just like other forms of replication.[104] The human gene PRNP codes for the major prion protein, PrP, and is subject to mutations that can give rise to disease-causing prions.
Beneficial mutations
Although mutations that cause changes in protein sequences can be harmful to an organism, on occasions the effect may be positive in a given environment. In this case, the mutation may enable the mutant organism to withstand particular environmental stresses better than wild-type organisms, or reproduce more quickly. In these cases a mutation will tend to become more common in a population through natural selection. Examples include the following:
HIV resistance: a specific 32 base pair deletion in human CCR5 (CCR5-Δ32) confers HIV resistance to homozygotes and delays AIDS onset in heterozygotes.[105] One possible explanation of the etiology of the relatively high frequency of CCR5-Δ32 in the European population is that it conferred resistance to the bubonic plague in mid-14th century Europe. People with this mutation were more likely to survive infection; thus its frequency in the population increased.[106] This theory could explain why this mutation is not found in Southern Africa, which remained untouched by bubonic plague. A newer theory suggests that the selective pressure on the CCR5 Delta 32 mutation was caused by smallpox instead of the bubonic plague.[107]
Malaria resistance: An example of a harmful mutation is sickle-cell disease, a blood disorder in which the body produces an abnormal type of the oxygen-carrying substance hemoglobin in the red blood cells. One-third of all indigenous inhabitants of Sub-Saharan Africa carry the allele, because, in areas where malaria is common, there is a survival value in carrying only a single sickle-cell allele (sickle cell trait).[108] Those with only one of the two alleles of the sickle-cell disease are more resistant to malaria, since the infestation of the malaria Plasmodium is halted by the sickling of the cells that it infests.
Antibiotic resistance: Practically all bacteria develop antibiotic resistance when exposed to antibiotics. In fact, bacterial populations already have such mutations that get selected under antibiotic selection.[109] Obviously, such mutations are only beneficial for the bacteria but not for those infected.
Lactase persistence. A mutation allowed humans to express the enzyme lactase after they are naturally weaned from breast milk, allowing adults to digest lactose, which is likely one of the most beneficial mutations in recent human evolution.[110]
Compensated pathogenic deviations
Compensated pathogenic deviations refer to amino acid residues in a protein sequence that are pathogenic in one species but are wild type residues in the functionally equivalent protein in another species. Although the amino acid residue is pathogenic in the first species, it is not so in the second species because its pathogenicity is compensated by one or more amino acid substitutions in the second species. The compensatory mutation can occur in the same protein or in another protein with which it interacts.[111]
It is critical to understand the effects of compensatory mutations in the context of fixed deleterious mutations due to the population fitness decreasing because of fixation.[112] Effective population size refers to a population that is reproducing.[113] An increase in this population size has been correlated with a decreased rate of genetic diversity.[113] The position of a population relative to the critical effect population size is essential to determine the effect deleterious alleles will have on fitness.[112] If the population is below the critical effective size fitness will decrease drastically, however if the population is above the critical effect size, fitness can increase regardless of deleterious mutations due to compensatory alleles.[112]
Compensatory mutations in RNA
As the function of a RNA molecule is dependent on its structure,[114] the structure of RNA molecules is evolutionarily conserved. Therefore, any mutation that alters the stable structure of RNA molecules must be compensated by other compensatory mutations. In the context of RNA, the sequence of the RNA can be considered as ' genotype' and the structure of the RNA can be considered as its 'phenotype'. Since RNAs have relatively simpler composition than proteins, the structure of RNA molecules can be computationally predicted with high degree of accuracy. Because of this convenience, compensatory mutations have been studied in computational simulations using RNA folding algorithms.[115][116]
Evolutionary mechanism of compensation
Compensatory mutations can be explained by the genetic phenomenon epistasis whereby the phenotypic effect of one mutation is dependent upon mutation(s) at other loci. While epistasis was originally conceived in the context of interaction between different genes, intragenic epistasis has also been studied recently.[117] Existence of compensated pathogenic deviations can be explained by 'sign epistasis', in which the effects of a deleterious mutation can be compensated by the presence of a epistatic mutation in another loci. For a given protein, a deleterious mutation (D) and a compensatory mutation (C) can be considered, where C can be in the same protein as D or in a different interacting protein depending on the context. The fitness effect of C itself could be neutral or somewhat deleterious such that it can still exist in the population, and the effect of D is deleterious to the extent that it cannot exist in the population. However, when C and D co-occur together, the combined fitness effect becomes neutral or positive.[111] Thus, compensatory mutations can bring novelty to proteins by forging new pathways of protein evolution : it allows individuals to travel from one fitness peak to another through the valleys of lower fitness.[117]
DePristo et al. 2005 outlined two models to explain the dynamics of compensatory pathogenic deviations (CPD).[118] In the first hypothesis P is a pathogenic amino acid mutation that and C is a neutral compensatory mutation.[118] Under these conditions, if the pathogenic mutation arises after a compensatory mutation, then P can become fixed in the population.[118] The second model of CPDs states that P and C are both deleterious mutations resulting in fitness valleys when mutations occur simultaneously.[118] Using publicly available, Ferrer-Costa et al. 2007 obtained compensatory mutations and human pathogenic mutation datasets that were characterized to determine what causes CPDs.[119] Results indicate that the structural constraints and the location in protein structure determine whether compensated mutations will occur.[119]
Experimental evidence of compensatory mutations
Experiment in bacteria
Lunzer et al.[120] tested the outcome of swapping divergent amino acids between two orthologous proteins of isopropymalate dehydrogenase (IMDH). They substituted 168 amino acids in Escherichia coli IMDH that are wild type residues in IMDH Pseudomonas aeruginosa. They found that over one third of these substitutions compromised IMDH enzymatic activity in the Escherichia coli genetic background. This demonstrated that identical amino acid states can result in different phenotypic states depending on the genetic background. Corrigan et al. 2011 demonstrated how staphylococcus aureus was able to grow normally without the presence of lipoteichoic acid due to compensatory mutations.[121] Whole genome sequencing results revealed that when Cyclic-di-AMP phosphodiesterase (GdpP) was disrupted in this bacterium, it compensated for the disappearance of the cell wall polymer, resulting in normal cell growth.[121]
Research has shown that bacteria can gain drug resistance through compensatory mutations that do not impede or having little effect on fitness.[122] Previous research from Gagneux et al. 2006 has found that laboratory grown M. tuberculosis strains with rifampicin resistance have reduced fitness, however drug resistant clinical strains of this pathogenic bacteria do not have reduced fitness.[123] Comas et al. 2012 used whole genome comparisons between clinical strains and lab derived mutants to determine the role and contribution of compensatory mutations in drug resistance to rifampicin.[122] Genome analysis reveal rifampicin resistant strains have a mutation in rpoA and rpoC.[122] A similar study investigated the bacterial fitness associated with compensatory mutations in rifampin resistant Escherichia coli.[124] Results obtained from this study demonstrate that drug resistance is linked to bacterial fitness as higher fitness costs are linked to greater transcription errors.[124]
Experiment in virus
Gong et al.[125] collected obtained genotype data of influenza nucleoprotein from different timelines and temporally ordered them according to their time of origin. Then they isolated 39 amino acid substitutions that occurred in different timelines and substituted them in a genetic background that approximated the ancestral genotype. They found that 3 of the 39 substitutions significantly reduced the fitness of the ancestral background. Compensatory mutations are new mutations that arise and have a positive or neutral impact on a populations fitness.[126] Previous research has shown that populations have can compensate detrimental mutations.[111][126][127] Burch and Chao tested Fisher's geometric model of adaptive evolution by testing whether bacteriophage φ6 evolves by small steps.[128] Their results showed that bacteriophage φ6 fitness declined rapidly and recovered in small steps .[128] Viral nucleoproteins have been shown to avoid cytotoxic T lymphocytes (CTLs) through arginine-to glycine substitutions.[129] This substitution mutations impacts the fitness of viral nucleoproteins, however compensatory co-mutations impede fitness declines and aid the virus to avoid recognition from CTLs.[129] Mutations can have three different effects; mutations can have deleterious effects, some increase fitness through compensatory mutations, and lastly mutations can be counterbalancing resulting in compensatory neutral mutations.[130][124][123]
See also
References
- Kimura, Motoo (1 July 1985). "The role of compensatory neutral mutations in molecular evolution". Journal of Genetics. 64 (1): 7–19. doi:10.1007/BF02923549. ISSN 0973-7731. S2CID 129866.
External links
- Jones S, Woolfson A, Partridge L (6 December 2007). "Genetic Mutation". In Our Time. BBC Radio 4. Retrieved 18 October 2015.
- Liou S (5 February 2011). "All About Mutations". HOPES. Huntington's Disease Outreach Project for Education at Stanford. Retrieved 18 October 2015.
- "Locus Specific Mutation Databases". Leiden, the Netherlands: Leiden University Medical Center. Retrieved 18 October 2015.
- "Welcome to the Mutalyzer website". Leiden, the Netherlands: Leiden University Medical Center. Retrieved 18 October 2015. – The Mutalyzer website.
https://en.wikipedia.org/wiki/Mutation#By_effect_on_function
Annexin A1, also known as lipocortin I, is a protein that is encoded by the ANXA1 gene in humans.[5]
https://en.wikipedia.org/wiki/Annexin_A1
Respiratory burst (or oxidative burst) is the rapid release of the reactive oxygen species (ROS), superoxide anion (O−
2) and hydrogen peroxide (H
2O
2), from different cell types.
This is usually utilised for mammalian immunological defence, but also plays a role in cell signalling. Respiratory burst is also implicated in the ovum of animals following fertilization. It may also occur in plant cells.
https://en.wikipedia.org/wiki/Respiratory_burst
In hematology, myelopoiesis in the broadest sense of the term is the production of bone marrow and of all cells that arise from it, namely, all blood cells.[1] In a narrower sense, myelopoiesis also refers specifically to the regulated formation of myeloid leukocytes (myelocytes), including eosinophilic granulocytes, basophilic granulocytes, neutrophilic granulocytes, and monocytes.[2]
The common myeloid progenitor can differentiate in the bone marrow into red blood cells and megakaryocytes (leading to platelets) as well as mast cells and myeloblasts, the latter leading to the myelocytic line (granulocytes) and to monocytes, macrophages, and dendritic cells of the innate immune system. The granulocytes, also called polymorphonuclear leukocytes because of their multilobed nuclei, are three short lived cell types including eosinophils, basophils, and neutrophils. A granulocyte differentiates into a distinct cell type by a process called granulopoiesis. In this process it first transforms from a common myeloblast (myeloid progenitor) to a common promyelocyte. This promyelocyte gives rise to a unique myelocyte that for the first time can be classified as an eosinophil, basophil, or neutrophil progenitor based on the histological staining affinity (eosinophilic, basophilic, or neutral granules).[3] The unique myelocyte next differentiates into a metamyelocyte and then a band cell, with a "C" shaped nucleus, before becoming a mature eosinophil, basophil, or neutrophil. Macrophages come from monoblast progenitors that differentiate into promonocytes, which mature into monocytes. Monocytes eventually enter the tissues and become macrophages.[citation needed]
https://en.wikipedia.org/wiki/Myelopoiesis
Granulopoiesis (or granulocytopoiesis) is a part of haematopoiesis, that leads to the production of granulocytes. A granulocyte, also referred to as a polymorphonuclear leukocyte (PMN), is a type of white blood cell that has multi lobed nuclei, usually containing three lobes, and has a significant amount of cytoplasmic granules within the cell.[1] Granulopoiesis takes place in the bone marrow.[2] It leads to the production of three types of mature granulocytes: neutrophils (most abundant, making up to 60% of all white blood cells), eosinophils (up to 4%) and basophils (up to 1%).[3]
https://en.wikipedia.org/wiki/Granulopoiesis
Hematopoietic stem cells (HSCs) are the stem cells[1] that give rise to other blood cells. This process is called haematopoiesis.[2] In vertebrates, the very first definitive HSCs arise from the ventral endothelial wall of the embryonic aorta within the (midgestational) aorta-gonad-mesonephros region, through a process known as endothelial-to-hematopoietic transition.[3][4] In adults, haematopoiesis occurs in the red bone marrow, in the core of most bones. The red bone marrow is derived from the layer of the embryo called the mesoderm.
https://en.wikipedia.org/wiki/Hematopoietic_stem_cell
| CFU-GEMM | |
|---|---|
 Red blood cells, white blood cells, and platelets are all derivatives of the CFU-GEMM cell. | |
| Details | |
| Gives rise to | Myeloid cells |
| Location | Bone marrow |
| Function | colony forming unit |
| Identifiers | |
| TH | H2.00.04.3.02008 |
| Anatomical terms of microanatomy | |
CFU-GEMM is a colony forming unit that generates myeloid cells. CFU-GEMM cells are the oligopotential progenitor cells[1][2] for myeloid cells; they are thus also called common myeloid progenitor cells or myeloid stem cells. "GEMM" stands for granulocyte, erythrocyte, monocyte, megakaryocyte.[3]
The common myeloid progenitor (CMP) and the common lymphoid progenitor (CLP) are the first branch of cell differentiation in hematopoiesis after the hemocytoblast (hematopoietic stem cell).
https://en.wikipedia.org/wiki/CFU-GEMM
In cell biology, a granule is a small particle.[1] It can be any structure barely visible by light microscopy. The term is most often used to describe a secretory vesicle.
https://en.wikipedia.org/wiki/Granule_(cell_biology)
An azurophilic granule is a cellular object readily stainable with a Romanowsky stain. In white blood cells and hyperchromatin, staining imparts a burgundy or merlot coloration. Neutrophils in particular are known for containing azurophils loaded with a wide variety of anti-microbial defensins that fuse with phagocytic vacuoles. Azurophils may contain myeloperoxidase, phospholipase A2, acid hydrolases, elastase, defensins, neutral serine proteases, bactericidal/permeability-increasing protein,[1] lysozyme, cathepsin G, proteinase 3, and proteoglycans.[citation needed]
Azurophil granules are also known as "primary granules".[2]
Furthermore, the term "azurophils" may refer to a unique type of cells, identified only in reptiles. These cells are similar in size to so-called heterophils with abundant cytoplasm that is finely to coarsely granular and may sometimes contain vacuoles. Granules may impart a purplish hue to the cytoplasm, particularly to the outer region. Occasionally, azurophils are observed with vacuolated cytoplasm.[3]
https://en.wikipedia.org/wiki/Azurophilic_granule
Specific granules are secretory vesicles found exclusively in cells of the immune system called granulocytes.
It is sometimes described as applying specifically to neutrophils,[1] and sometimes the term is applied to other types of cells.[2]
These granules store a mixture of cytotoxic molecules, including many enzymes and antimicrobial peptides, that are released by a process called degranulation following activation of the granulocyte by an immune stimulus.
Specific granules are also known as "secondary granules".[3]
Contents
Examples of cytotoxic molecule stored by specific granules in different granulocytes include:
- Neutrophil: alkaline phosphatase, lactoferrin, lysozyme, NADPH oxidase
- Eosinophil: cathepsin, major basic protein
- Basophil: heparin, histamine (not directly cytotoxic)
Clinical significance
A specific granule deficiency can be associated with CEBPE.[4]
https://en.wikipedia.org/wiki/Specific_granule
| Dense granule | |
|---|---|
 Dense granules shown in a platelet | |
| Details | |
| Identifiers | |
| Latin | granulum delta |
| TH | H2.00.04.1.03006 |
| Anatomical terminology | |
Dense granules (also known as dense bodies or delta granules) are specialized secretory organelles. Dense granules are found only in platelets and are smaller than alpha granules.[1] The origin of these dense granules is still unknown, however, it is thought that may come from the mechanism involving the endocytotic pathway.[2] Dense granules are a sub group of lysosome-related organelles (LRO). There are about three to eight of these in a normal human platelet.[3]
https://en.wikipedia.org/wiki/Dense_granule
The parasitophorous vacuole (PV) is a structure produced by apicomplexan parasites in the cells of its host. The PV allows the parasite to develop while protected from the phagolysosomes of the host cell.[1]
The PV is a bubble-like compartment made of plasma membrane; the compartment contains cytoplasm and the parasite. The PV allows the parasite to exist and grow within the cell while protecting the parasite from the host cell defense mechanisms. The PV prevents the acidification of the compartment, the mechanism by which the lysosomes of the host cell would normally destroy an invading parasite.[1] Parasites that form a parasitophorous vacuole as part of their infection process include Plasmodium falciparum, which causes malaria and Toxoplasma gondii, which causes toxoplasmosis.
The parasitophorous vacuole is formed during cell invasion, when the parasite uses part of the membrane of the host cell to form a parasitophorous vacuolar membrane (PVM). The PVM surrounds the intracellular parasite, creating a separate bubble of cytoplasm-filled plasma membrane within the host cell. The rhoptry and the microneme, special secretory organelles found in apicomplexan parasites, play a major role in the formation of the vacuole.[2][3] The PVM is extensively re-modelled by parasitic proteins.[4] One theory is that the microneme works with the rhoptry and the rhoptry secretes proteins to create the PVM, while the microneme binds to the surface of red blood cells, allowing the parasite to more easily enter into the cell.[5]
The PV is not a true vacuole, but resembles one under the microscope.[5]
https://en.wikipedia.org/wiki/Parasitophorous_vacuole
Coagulation cascade
The coagulation cascade of secondary hemostasis has two initial pathways which lead to fibrin formation. These are the contact activation pathway (also known as the intrinsic pathway), and the tissue factor pathway (also known as the extrinsic pathway), which both lead to the same fundamental reactions that produce fibrin. It was previously thought that the two pathways of coagulation cascade were of equal importance, but it is now known that the primary pathway for the initiation of blood coagulation is the tissue factor (extrinsic) pathway. The pathways are a series of reactions, in which a zymogen (inactive enzyme precursor) of a serine protease and its glycoprotein co-factor are activated to become active components that then catalyze the next reaction in the cascade, ultimately resulting in cross-linked fibrin. Coagulation factors are generally indicated by Roman numerals, with a lowercase a appended to indicate an active form.[7]
The coagulation factors are generally enzymes called serine proteases, which act by cleaving downstream proteins. The exceptions are tissue factor, FV, FVIII, FXIII.[8] Tissue factor, FV and FVIII are glycoproteins, and Factor XIII is a transglutaminase.[7] The coagulation factors circulate as inactive zymogens. The coagulation cascade is therefore classically divided into three pathways. The tissue factor and contact activation pathways both activate the "final common pathway" of factor X, thrombin and fibrin.[9]
Tissue factor pathway (extrinsic)
The main role of the tissue factor (TF) pathway is to generate a "thrombin burst", a process by which thrombin, the most important constituent of the coagulation cascade in terms of its feedback activation roles is released very rapidly. FVIIa circulates in a higher amount than any other activated coagulation factor. The process includes the following steps:[7]
- Following damage to the blood vessel, FVII leaves the circulation and comes into contact with tissue factor expressed on tissue-factor-bearing cells (stromal fibroblasts and leukocytes), forming an activated complex (TF-FVIIa).
- TF-FVIIa activates FIX and FX.
- FVII is itself activated by thrombin, FXIa, FXII, and FXa.
- The activation of FX (to form FXa) by TF-FVIIa is almost immediately inhibited by tissue factor pathway inhibitor (TFPI).
- FXa and its co-factor FVa form the prothrombinase complex, which activates prothrombin to thrombin.
- Thrombin then activates other components of the coagulation cascade, including FV and FVIII (which forms a complex with FIX), and activates and releases FVIII from being bound to vWF.
- FVIIIa is the co-factor of FIXa, and together they form the "tenase" complex, which activates FX; and so the cycle continues. ("Tenase" is a contraction of "ten" and the suffix "-ase" used for enzymes.)
Contact activation pathway (intrinsic)
The contact activation pathway begins with formation of the primary complex on collagen by high-molecular-weight kininogen (HMWK), prekallikrein, and FXII (Hageman factor). Prekallikrein is converted to kallikrein and FXII becomes FXIIa. FXIIa converts FXI into FXIa. Factor XIa activates FIX, which with its co-factor FVIIIa form the tenase complex, which activates FX to FXa. The minor role that the contact activation pathway has in initiating clot formation can be illustrated by the fact that individuals with severe deficiencies of FXII, HMWK, and prekallikrein do not have a bleeding disorder. Instead, contact activation system seems to be more involved in inflammation,[7] and innate immunity.[10] Despite this, interference with the pathway may confer protection against thrombosis without a significant bleeding risk.[10]
Final common pathway
The division of coagulation in two pathways is arbitrary, originating from laboratory tests in which clotting times were measured either after the clotting was initiated by glass, the intrinsic pathway; or clotting was initiated by thromboplastin (a mix of tissue factor and phospholipids), the extrinsic pathway.[citation needed]
Further, the final common pathway scheme implies that prothrombin is converted to thrombin only when acted upon by the intrinsic or extrinsic pathways, which is an oversimplification. In fact, thrombin is generated by activated platelets at the initiation of the platelet plug, which in turn promotes more platelet activation.[citation needed]
Thrombin functions not only to convert fibrinogen to fibrin, it also activates Factors VIII and V and their inhibitor protein C (in the presence of thrombomodulin). By activating Factor XIII, covalent bonds are formed that crosslink the fibrin polymers that form from activated monomers.[7] This stabilizes the fibrin network.
The coagulation cascade is maintained in a prothrombotic state by the continued activation of FVIII and FIX to form the tenase complex until it is down-regulated by the anticoagulant pathways.[7]
Cell-based scheme of coagulation
A newer model of coagulation mechanism explains the intricate combination of cellular and biochemical events that occur during the coagulation process in vivo. Along with the procoagulant and anticoagulant plasma proteins, normal physiologic coagulation requires the presence of two cell types for formation of coagulation complexes: cells that express tissue factor (usually extravascular) and platelets.[citation needed]
The coagulation process occurs in two phases. First is the initiation phase, which occurs in tissue-factor-expressing cells. This is followed by the propagation phase, which occurs on activated platelets. The initiation phase, mediated by the tissue factor exposure, proceeds via the classic extrinsic pathway and contributes to about 5% of thrombin production. The amplified production of thrombin occurs via the classic intrinsic pathway in the propagation phase; about 95% of thrombin generated will be during this second phase.[11]
Cofactors
Various substances are required for the proper functioning of the coagulation cascade:
Calcium and phospholipids
Calcium and phospholipids (constituents of platelet membrane) are required for the tenase and prothrombinase complexes to function.[12] Calcium mediates the binding of the complexes via the terminal gamma-carboxy residues on Factor Xa and Factor IXa to the phospholipid surfaces expressed by platelets, as well as procoagulant microparticles or microvesicles shed from them.[13] Calcium is also required at other points in the coagulation cascade. Calcium ions play a major role in the regulation of coagulation cascade that is paramount in the maintenance of hemostasis. Other than platelet activation, calcium ions are responsible for complete activation of several coagulation factors, including coagulation Factor XIII.[14]
Vitamin K
Vitamin K is an essential factor to a hepatic gamma-glutamyl carboxylase that adds a carboxyl group to glutamic acid residues on factors II, VII, IX and X, as well as Protein S, Protein C and Protein Z. In adding the gamma-carboxyl group to glutamate residues on the immature clotting factors, Vitamin K is itself oxidized. Another enzyme, Vitamin K epoxide reductase (VKORC), reduces vitamin K back to its active form. Vitamin K epoxide reductase is pharmacologically important as a target of anticoagulant drugs warfarin and related coumarins such as acenocoumarol, phenprocoumon, and dicumarol. These drugs create a deficiency of reduced vitamin K by blocking VKORC, thereby inhibiting maturation of clotting factors. Vitamin K deficiency from other causes (e.g., in malabsorption) or impaired vitamin K metabolism in disease (e.g., in liver failure) lead to the formation of PIVKAs (proteins formed in vitamin K absence), which are partially or totally non-gamma carboxylated, affecting the coagulation factors' ability to bind to phospholipid.[citation needed]
Regulators
Five mechanisms keep platelet activation and the coagulation cascade in check.[citation needed] Abnormalities can lead to an increased tendency toward thrombosis:
Protein C
Protein C is a major physiological anticoagulant. It is a vitamin K-dependent serine protease enzyme that is activated by thrombin into activated protein C (APC). Protein C is activated in a sequence that starts with Protein C and thrombin binding to a cell surface protein thrombomodulin. Thrombomodulin binds these proteins in such a way that it activates Protein C. The activated form, along with protein S and a phospholipid as cofactors, degrades FVa and FVIIIa. Quantitative or qualitative deficiency of either (protein C or protein S) may lead to thrombophilia (a tendency to develop thrombosis). Impaired action of Protein C (activated Protein C resistance), for example by having the "Leiden" variant of Factor V or high levels of FVIII, also may lead to a thrombotic tendency.[citation needed]
Antithrombin
Antithrombin is a serine protease inhibitor (serpin) that degrades the serine proteases: thrombin, FIXa, FXa, FXIa, and FXIIa. It is constantly active, but its adhesion to these factors is increased by the presence of heparan sulfate (a glycosaminoglycan) or the administration of heparins (different heparinoids increase affinity to FXa, thrombin, or both). Quantitative or qualitative deficiency of antithrombin (inborn or acquired, e.g., in proteinuria) leads to thrombophilia.[citation needed]
Tissue factor pathway inhibitor (TFPI)
Tissue factor pathway inhibitor (TFPI) limits the action of tissue factor (TF). It also inhibits excessive TF-mediated activation of FVII and FX.[citation needed]
Plasmin
Plasmin is generated by proteolytic cleavage of plasminogen, a plasma protein synthesized in the liver. This cleavage is catalyzed by tissue plasminogen activator (t-PA), which is synthesized and secreted by endothelium. Plasmin proteolytically cleaves fibrin into fibrin degradation products that inhibit excessive fibrin formation.[citation needed]
Prostacyclin
Prostacyclin (PGI2) is released by endothelium and activates platelet Gs protein-linked receptors. This, in turn, activates adenylyl cyclase, which synthesizes cAMP. cAMP inhibits platelet activation by decreasing cytosolic levels of calcium and, by doing so, inhibits the release of granules that would lead to activation of additional platelets and the coagulation cascade.[15]
Fibrinolysis
Eventually, blood clots are reorganized and resorbed by a process termed fibrinolysis. The main enzyme responsible for this process (plasmin) is regulated by various activators and inhibitors.[15]
Role in immune system
The coagulation system overlaps with the immune system. Coagulation can physically trap invading microbes in blood clots. Also, some products of the coagulation system can contribute to the innate immune system by their ability to increase vascular permeability and act as chemotactic agents for phagocytic cells. In addition, some of the products of the coagulation system are directly antimicrobial. For example, beta-lysine, an amino acid produced by platelets during coagulation, can cause lysis of many Gram-positive bacteria by acting as a cationic detergent.[16] Many acute-phase proteins of inflammation are involved in the coagulation system. In addition, pathogenic bacteria may secrete agents that alter the coagulation system, e.g. coagulase and streptokinase.[citation needed]
https://en.wikipedia.org/wiki/Coagulation#The_coagulation_cascade
Heřmanský–Pudlák syndrome (often written Hermansky–Pudlak syndrome or abbreviated HPS) is an extremely rare autosomal recessive[1] disorder which results in oculocutaneous albinism (decreased pigmentation), bleeding problems due to a platelet abnormality (platelet storage pool defect), and storage of an abnormal fat-protein compound (lysosomal accumulation of ceroid lipofuscin). It is thought to affect around 1 in 500,000 people worldwide, with a significantly higher occurrence in Puerto Ricans, with a prevalence of 1 in 1800.[2] Many of the clinical research studies on the disease have been conducted in Puerto Rico.
There are eight classic forms of the disorder, based on the genetic mutation from which the disorder stems.[3]
It is named for František Heřmanský (1916–1980) and Pavel Pudlák (1927–1993).[4][5][6]
https://en.wikipedia.org/wiki/Hermansky%E2%80%93Pudlak_syndrome
| Band cell | |
|---|---|
| Details | |
| Precursor | metamyelocyte |
| Gives rise to | Granulocyte |
| Identifiers | |
| TH | H2.00.04.3.04011 |
| FMA | 86471 |
| Anatomical terms of microanatomy | |
A band cell (also called band neutrophil, band form or stab cell) is a cell undergoing granulopoiesis, derived from a metamyelocyte, and leading to a mature granulocyte.
It is characterized by having a curved but not lobular nucleus.[1]
The term "band cell" implies a granulocytic lineage (e.g., neutrophils).[2]
https://en.wikipedia.org/wiki/Band_cell
Monocytopoiesis is the process which leads to the production of monocytes (and, subsequently, macrophages).
It can be induced by macrophage colony-stimulating factor.[citation needed]
It is a component of myelopoiesis.[1]
https://en.wikipedia.org/wiki/Monocytopoiesis
Extramedullary hematopoiesis (EMH or sometimes EH[1]) refers to hematopoiesis occurring outside of the medulla of the bone (bone marrow).[2] It can be physiologic or pathologic.
Physiologic EMH occurs during embryonic and fetal development; during this time the main site of fetal hematopoiesis are liver and the spleen.
Pathologic EMH can occur during adulthood when physiologic hematopoiesis can't work properly in the bone marrow and the hematopoietic stem cells (HSC) have to migrate to other tissues in order to continue with the formation of blood cellular components. Pathologic EMH can be caused by myelofibrosis,[3] thalassemias or disorders caused in the hematopoietic system.
https://en.wikipedia.org/wiki/Extramedullary_hematopoiesis
| Monoblast | |
|---|---|
 Monoblast | |
| Details | |
| System | Immune system |
| Location | Bone marrow |
| Function | A precursor monocyte |
| Identifiers | |
| TH | H2.00.04.3.08002 |
| FMA | 83553 |
| Anatomical terms of microanatomy | |
Monoblasts are the committed progenitor cells that differentiated from a committed macrophage or dendritic cell precursor (MDP[1]) in the process of hematopoiesis. They are the first developmental stage in the monocyte series leading to a macrophage.[2] Their myeloid cell fate is induced by the concentration of cytokines they are surrounded by during development. These cytokines induce the activation of transcription factors which push completion of the monoblast's myeloid cell fate. Monoblasts are normally found in bone marrow and do not appear in the normal peripheral blood.[3] They mature into monocytes which, in turn, develop into macrophages.[4] They then are seen as macrophages in the normal peripheral blood and many different tissues of the body. Macrophages can produce a variety of effector molecules that initiate local, systemic inflammatory responses. These monoblast differentiated cells are equipped to fight off foreign invaders using pattern recognition receptors to detect antigen as part of the innate immune response. [1]
https://en.wikipedia.org/wiki/Monoblast
A foreign-body giant cell is a collection of fused macrophages (giant cell) which are generated in response to the presence of a large foreign body. This is particularly evident with catheters, parasites, or biomaterials that are inserted into the body for replacement or regeneration of diseased or damaged tissues.[1][2] Foreign body giant cells are also produced to digest foreign material that is too large for phagocytosis.[3] The inflammatory process that creates these cells often leads to a foreign body granuloma.
The human body goes through several steps when exposed to foreign biomaterial including acute and chronic inflammation, and formation of new tissue and a fibrous capsule along the surface of the implantation.[1] Foreign body reactions, which are a type of chronic inflammation, are characterized by the presence of macrophages, monocytes, and foreign-body giant cells (FBGCs).[1][4] The response of the foreign body reaction determines how compatible the implanted material will be in the body, and the members of the foreign body reaction, including the FBGC's, remain along the surface of the biomaterial for its lifetime in the body.[1]
Foreign body giant cells are formed through signaling from IL-4 and IL-13, and may fuse to produce a multinucleated cell with up to 200 nuclei within its cytoplasm.[5]
https://en.wikipedia.org/wiki/Foreign-body_giant_cell
| Foreign body granuloma | |
|---|---|
 | |
| Transvaginal ultrasonography showing a foreign body granuloma at right as a hypoechoic (dark) area around a perforated intrauterine device. The uterus is at left. | |
| Specialty | Dermatology |
A foreign body reaction (FBR) is a typical tissue response to a foreign body within biological tissue.[1] It usually includes the formation of a foreign body granuloma.[2] Tissue-encapsulation of an implant is an example, as is inflammation around a splinter.[3] Foreign body granuloma formation consists of protein adsorption, macrophages, multinucleated foreign body giant cells (macrophage fusion), fibroblasts, and angiogenesis. It has also been proposed that the mechanical property of the interface between an implant and its surrounding tissues is critical for the host response.[4]
In the long term, the foreign body reaction results in encapsulation of the foreign body within a calcified shell. For example, a lithopedion is a rare phenomenon which occurs most commonly when a fetus dies during an abdominal pregnancy,[5] is too large to be reabsorbed by the body, and calcifies.
https://en.wikipedia.org/wiki/Foreign_body_reaction
| Reticulocyte | |
|---|---|
 Reticulocytes | |
 Erythrocytes (mature cells) | |
| Details | |
| Gives rise to | Red blood cells |
| Location | Bone marrow (most), blood (some) |
| Identifiers | |
| Latin | reticulocytus |
| MeSH | D012156 |
| TH | H2.00.04.1.01007 |
| FMA | 66785 |
| Anatomical terms of microanatomy | |
Reticulocytes are immature red blood cells (RBCs). In the process of erythropoiesis (red blood cell formation), reticulocytes develop and mature in the bone marrow and then circulate for about a day in the blood stream before developing into mature red blood cells. Like mature red blood cells, in mammals, reticulocytes do not have a cell nucleus. They are called reticulocytes because of a reticular (mesh-like) network of ribosomal RNA that becomes visible under a microscope with certain stains such as new methylene blue and Romanowsky stain.
https://en.wikipedia.org/wiki/Reticulocyte
A nucleated red blood cell (NRBC), also known by several other names, is a red blood cell that contains a cell nucleus. Almost all vertebrate organisms have hemoglobin-containing cells in their blood, and with the exception of mammals, all of these red blood cells are nucleated.[1] In mammals, NRBCs occur in normal development as precursors to mature red blood cells in erythropoiesis, the process by which the body produces red blood cells. NRBCs are normally found in the bone marrow of humans of all ages and in the blood of fetuses and newborn infants.[2][3] After infancy, RBCs normally contain a nucleus only during the very early stages of the cell's life, and the nucleus is ejected as a normal part of cellular differentiation before the cell is released into the bloodstream. Thus, if NRBCs are identified on an adult's complete blood count or peripheral blood smear, it suggests that there is a very high demand for the bone marrow to produce RBCs, and immature RBCs are being released into circulation. Possible pathologic causes include anemia, myelofibrosis, thalassemia, miliary tuberculosis, cancers involving bone marrow (myelomas, leukemias, lymphomas), and in chronic hypoxemia.[4]
https://en.wikipedia.org/wiki/Nucleated_red_blood_cell
Leukocyte extravasation (also commonly known as leukocyte adhesion cascade or diapedesis – the passage of cells through the intact vessel wall) is the movement of leukocytes out of the circulatory system and towards the site of tissue damage or infection. This process forms part of the innate immune response, involving the recruitment of non-specific leukocytes. Monocytes also use this process in the absence of infection or tissue damage during their development into macrophages.
https://en.wikipedia.org/wiki/Leukocyte_extravasation
Chemoattractants and chemorepellents
Chemoattractants and chemorepellents are inorganic or organic substances possessing chemotaxis-inducer effect in motile cells. These chemotactic ligands create chemical concentration gradients that organisms, prokaryotic and eukaryotic, move toward or away from, respectively.[34]
Effects of chemoattractants are elicited via chemoreceptors such as methyl-accepting chemotaxis proteins (MCP).[35] MCPs in E.coli include Tar, Tsr, Trg and Tap.[36] Chemoattracttants to Trg include ribose and galactose with phenol as a chemorepellent. Tap and Tsr recognize dipeptides and serine as chemoattractants, respectively.[36]
Chemoattractants or chemorepellents bind MCPs at its extracellular domain; an intracellular signaling domain relays the changes in concentration of these chemotactic ligands to downstream proteins like that of CheA which then relays this signal to flagellar motors via phosphorylated CheY (CheY-P).[35] CheY-P can then control flagellar rotation influencing the direction of cell motility.[35]
For E.coli, S. meliloti, and R. spheroids, the binding of chemoattractants to MCPs inhibit CheA and therefore CheY-P activity, resulting in smooth runs, but for B. substilis, CheA activity increases.[35] Methylation events in E.coli cause MCPs to have lower affinity to chemoattractants which causes increased activity of CheA and CheY-P resulting in tumbles.[35] In this way cells are able to adapt to the immediate chemoattractant concentration and detect further changes to modulate cell motility.[35]
Chemoattractants in eukaryotes are well characterized for immune cells. Formyl peptides, such as fMLF, attract leukocytes such as neutrophils and macrophages, causing movement toward infection sites.[37] Non-acylated methioninyl peptides do not act as chemoattractants to neutrophils and macrophages.[37] Leukocytes also move toward chemoattractants C5a, a complement component, and pathogen-specific ligands on bacteria.[37]
Mechanisms concerning chemorepellents are less known than chemoattractants. Although chemorepellents work to confer an avoidance response in organisms, Tetrahymena thermophila adapt to a chemorepellent, Netrin-1 peptide, within 10 minutes of exposure; however, exposure to chemorepellents such as GTP, PACAP-38, and nociceptin show no such adaptations.[38] GTP and ATP are chemorepellents in micro-molar concentrations to both Tetrahymena and Paramecium. These organisms avoid these molecules by producing avoiding reactions to re-orient themselves away from the gradient.[39]
https://en.wikipedia.org/wiki/Chemotaxis#Chemoattractants_and_chemorepellents
Leukotrienes are a family of eicosanoid inflammatory mediators produced in leukocytes by the oxidation of arachidonic acid (AA) and the essential fatty acid eicosapentaenoic acid (EPA) by the enzyme arachidonate 5-lipoxygenase.[1][2][3]
Leukotrienes use lipid signaling to convey information to either the cell producing them (autocrine signaling) or neighboring cells (paracrine signaling) in order to regulate immune responses. The production of leukotrienes is usually accompanied by the production of histamine and prostaglandins, which also act as inflammatory mediators.[4]
One of their roles (specifically, leukotriene D4) is to trigger contractions in the smooth muscles lining the bronchioles; their overproduction is a major cause of inflammation in asthma and allergic rhinitis.[5] Leukotriene antagonists are used to treat these disorders by inhibiting the production or activity of leukotrienes.[6]
History and name
The name leukotriene, introduced by Swedish biochemist Bengt Samuelsson in 1979, comes from the words leukocyte and triene (indicating the compound's three conjugated double bonds). What would be later named leukotriene C, "slow reaction smooth muscle-stimulating substance" (SRS) was originally described between 1938 and 1940 by Feldberg and Kellaway.[7] [8][9] The researchers isolated SRS from lung tissue after a prolonged period following exposure to snake venom and histamine.[10]
https://en.wikipedia.org/wiki/Leukotriene
| RELA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||
| |||||||||||||
| |||||||
| Identifiers | |||||||
|---|---|---|---|---|---|---|---|
| Aliases | RELA, NFKB3, p65, RELA proto-oncogene, NF-kB subunit, CMCU | ||||||
| External IDs | OMIM: 164014 MGI: 103290 HomoloGene: 32064 GeneCards: RELA | ||||||
| |||||||
| |||||||
| |||||||
| |||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Transcription factor p65 also known as nuclear factor NF-kappa-B p65 subunit is a protein that in humans is encoded by the RELA gene.[5]
RELA, also known as p65, is a REL-associated protein involved in NF-κB heterodimer formation, nuclear translocation and activation[citation needed]. NF-κB is an essential transcription factor complex involved in all types of cellular processes, including cellular metabolism, chemotaxis, etc. Phosphorylation and acetylation of RELA are crucial post-translational modifications required for NF-κB activation. RELA has also been shown to modulate immune responses, and activation of RELA is positively associated with multiple types of cancer.
https://en.wikipedia.org/wiki/RELA
An immunotoxin is an artificial protein consisting of a targeting portion linked to a toxin. When the protein binds to that cell, it is taken in through endocytosis, and the toxin kills the cell.[1] They are used for the treatment of some kinds of cancer and a few viral infections.
Design
These chimeric proteins are usually made of a modified antibody or antibody fragment, attached to a fragment of a toxin. The targeting portion is composed of the Fab portion of an antibody that targets a specific cell type.[2] The toxin is usually an AB toxin, a cytotoxic protein derived from a bacterial or plant protein, from which the natural binding domain has been removed so that the Fv directs the toxin to the antigen on the target cell.[1]
Sometimes recombinant fusion proteins containing a toxin and a growth factor are also referred to as recombinant immunotoxins, although they do not contain an antibody fragment. A more specific name for this latter kind of protein is recombinant fusion toxin.
Production
They were originally produced by attaching the antibody to the toxin using a chemical linker.[citation needed] They are now made using recombinant DNA techniques, are produced in bacteria like E. coli called recombinant immunotoxins.[3]
Function
The antibody (or other targeting moiety) binds to an antigen on the target cell and the toxin then enters and kills the cell.
Clinical application
The best clinical success has been achieved in treating patients with refractory hairy cell leukemia. These patients were treated with the recombinant immunotoxin which targets the CD22 cell surface receptor on these leukemic cells. In two uncontrolled clinical studies, about half of participants achieved a complete response after BL22 treatment.[4] This therapeutic has been superseded by HA22, a slightly modified version.
A recent Phase I study of Resimmune found an 89% response rate in a subgroup of nine patients with cutaneous T cell lymphoma.[5] This subgroup was Stage IB-IIB with mSWAT scores of less than 50. The complete response rate was 50% (two of which are over 72 months duration and could represent cures).
See also
- Denileukin diftitox, an immunotoxin used to treat T cell leukemias
- Resimmune, or A-dmDT390-bisFv(UCHT1), an experimental anti-T cell immunotoxin in clinical trials to treat cutaneous T-cell lymphoma and metastatic melanoma.
- Anti-CD22 immunotoxins, being tested in hairy cell leukemia
- Antibody-drug conjugates, most, if not all, are immunotoxins but use whole antibodies and non-protein drugs
- Immunopharmacology and Immunotoxicology (medical journal)
References
- [1] Angimmune: Clinical Trials: Identification of a Cutaneous T-Cell Lymphoma (CTCL) Subgroup Experiencing a High Treatment Response Rate: Paragraph 1
External links
- immunotoxins at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
https://en.wikipedia.org/wiki/Immunotoxin
National Academy of Medicine
Starting in the early 1990s, the federal government directed the Institute of Medicine (IOM), now known as the National Academy of Medicine, to issue reports every 2 years on the health effects of Agent Orange and similar herbicides. First published in 1994 and titled Veterans and Agent Orange, the IOM reports assess the risk of both cancer and non-cancer health effects. Each health effect is categorized by evidence of association based on available research data.[3] The last update was published in 2016, entitled "Veterans and Agent Orange: Update 2014."
The report shows sufficient evidence of an association with soft tissue sarcoma; non-Hodgkin lymphoma (NHL); Hodgkin disease; Chronic lymphocytic leukemia (CLL); including hairy cell leukemia and other chronic B-cell leukemias. Limited or suggested evidence of an association was linked with respiratory cancers (lung, bronchus, trachea, larynx); prostate cancer; multiple myeloma; and bladder cancer. Numerous other cancers were determined to have inadequate or insufficient evidence of links to Agent Orange.
The National Academy of Medicine has repeatedly concluded that any evidence suggestive of an association between Agent Orange and prostate cancer is, "limited because chance, bias, and confounding could not be ruled out with confidence."[67]
At the request of the Veterans Administration, the Institute Of Medicine evaluated whether service in these C-123 aircraft could have plausibly exposed soldiers and been detrimental to their health. Their report "Post-Vietnam Dioxin Exposure in Agent Orange-Contaminated C-123 Aircraft" confirmed it.[68]
U.S. Public Health Service
Publications by the United States Public Health Service have shown that Vietnam veterans, overall, have increased rates of cancer, and nerve, digestive, skin, and respiratory disorders. The Centers for Disease Control and Prevention notes that in particular, there are higher rates of acute/chronic leukemia, Hodgkin's lymphoma and non-Hodgkin's lymphoma, throat cancer, prostate cancer, lung cancer, colon cancer, Ischemic heart disease, soft tissue sarcoma, and liver cancer.[69][70] With the exception of liver cancer, these are the same conditions the U.S. Veterans Administration has determined may be associated with exposure to Agent Orange/dioxin and are on the list of conditions eligible for compensation and treatment.[70][71]
Military personnel who were involved in storage, mixture and transportation (including aircraft mechanics), and actual use of the chemicals were probably among those who received the heaviest exposures.[72] Military members who served on Okinawa also claim to have been exposed to the chemical, but there is no verifiable evidence to corroborate these claims.[73]
Some studies have suggested that veterans exposed to Agent Orange may be more at risk of developing prostate cancer[74] and potentially more than twice as likely to develop higher-grade, more lethal prostate cancers.[75] However, a critical analysis of these studies and 35 others consistently found that there was no significant increase in prostate cancer incidence or mortality in those exposed to Agent Orange or 2,3,7,8-tetracholorodibenzo-p-dioxin.[76]
https://en.wikipedia.org/wiki/Agent_Orange
https://en.wikipedia.org/wiki/Philosopher%27s_stone
https://en.wikipedia.org/wiki/Thomas_%26_Friends
https://en.wikipedia.org/wiki/Bob_the_Builder
Antibody-drug conjugates or ADCs are a class of biopharmaceutical drugs designed as a targeted therapy for treating cancer.[1] Unlike chemotherapy, ADCs are intended to target and kill tumor cells while sparing healthy cells. As of 2019, some 56 pharmaceutical companies were developing ADCs.[2]
ADCs are complex molecules composed of an antibody linked to a biologically active cytotoxic (anticancer) payload or drug.[3] Antibody-drug conjugates are an example of bioconjugates and immunoconjugates.
ADCs combine the targeting properties of monoclonal antibodies with the cancer-killing capabilities of cytotoxic drugs, designed to discriminate between healthy and diseased tissue.[4][5]
https://en.wikipedia.org/wiki/Antibody-drug_conjugate
The primate T-lymphotropic viruses (PTLVs) are a group of retroviruses that infect primates, using their lymphocytes to reproduce. The ones that infect humans are known as human T-lymphotropic virus (HTLV), and the ones that infect Old World monkeys are called simian T-lymphotropic viruses (STLVs). PTLVs are named for their ability to cause adult T-cell leukemia/lymphoma, but in the case of HTLV-1 it can also cause a demyelinating disease called tropical spastic paraparesis.[2] On the other hand, newer PTLVs are simply placed into the group by similarity and their connection to human disease remains unclear.[1]
HTLVs have evolved from STLVs by interspecies transmission. Within each species of PTLV, the HTLV is more similar to its cognate STLV than to the other HTLVs.[1] There are currently three species of PTLVs recognized by the ICTV (P/H/STLV-1, -2, -3), plus two that are reported but unrecognized (HTLV-4, STLV-5).[1] The first known, and still most medically important PTLV is HTLV-1, discovered in 1980.[3]
HTLVs belong to the genus Deltaretrovirus. The only other recognized species in the genus is Bovine leukemia virus, an economically-important cattle pathogen. As its name suggests, this virus causes leukemia in cows.[4]
https://en.wikipedia.org/wiki/Primate_T-lymphotropic_virus
Interferon alfacon-1 is a recombinant synthetic type I interferon used for the treatment of hairy cell leukemia, malignant melanoma and AIDS-related Kaposi's sarcoma.[1]
https://en.wikipedia.org/wiki/Interferon_alfacon-1
| Pyoderma gangrenosum | |
|---|---|
 | |
| Pyoderma gangrenosum on the leg of a person with ulcerative colitis. | |
| Specialty | Dermatology |
| Usual onset | 40s or 50s[1] |
| Treatment | Corticosteroids, ciclosporin, infliximab, canakinumab[2] |
Pyoderma gangrenosum is a rare, inflammatory skin disease where painful pustules or nodules become ulcers that progressively grow.[3] Pyoderma gangrenosum is not infectious.[3]
Treatments may include corticosteroids, ciclosporin, infliximab, or canakinumab.[2]
The disease was identified in 1930. It affects approximately 1 person in 100,000 in the population. Though it can affect people of any age, it mostly affects people in their 40s and 50s.[1]
https://en.wikipedia.org/wiki/Pyoderma_gangrenosum
| Interferon Type I (α/β/δ...) | |||
|---|---|---|---|
 | |||
| Identifiers | |||
| Symbol | Interferons | ||
| Pfam | PF00143 | ||
| InterPro | IPR000471 | ||
| SMART | SM00076 | ||
| PROSITE | PDOC00225 | ||
| CATH | 1au1 | ||
| SCOP2 | 1au1 / SCOPe / SUPFAM | ||
| CDD | cd00095 | ||
|
The type-I interferons (IFN) are cytokines which play essential roles in inflammation, immunoregulation, tumor cells recognition, and T-cell responses. In the human genome, a cluster of thirteen functional IFN genes is located at the 9p21.3 cytoband over approximately 400 kb including coding genes for IFNα (IFNA1, IFNA2, IFNA4, IFNA5, IFNA6, IFNA7, IFNA8, IFNA10, IFNA13, IFNA14, IFNA16, IFNA17 and IFNA21), IFNω (IFNW1), IFNɛ (IFNE), IFNк (IFNK) and IFNβ (IFNB1), plus 11 IFN pseudogenes.[1]
Interferons bind to interferon receptors. All type I IFNs bind to a specific cell surface receptor complex known as the IFN-α receptor (IFNAR) that consists of IFNAR1 and IFNAR2 chains.
Type I IFNs are found in all mammals, and homologous (similar) molecules have been found in birds, reptiles, amphibians and fish species.[2][3]
https://en.wikipedia.org/wiki/Interferon_type_I
| Polyarteritis nodosa | |
|---|---|
| Other names | Panarteritis nodosa,[1] Periarteritis nodosa,[1] Kussmaul disease, or Kussmaul-Maier disease,[2] |
 | |
| Polyarteritis nodosa: Macroscopic specimen of the heart with abundant adipose tissue and nodular thickened coronary vessels | |
| Specialty | Immunology, rheumatology |
Polyarteritis nodosa (PAN) is a systemic necrotizing inflammation of blood vessels (vasculitis) affecting medium-sized muscular arteries, typically involving the arteries of the kidneys and other internal organs but generally sparing the lungs' circulation.[3] Small aneurysms are strung like the beads of a rosary,[4] therefore making this "rosary sign" an important diagnostic feature of the vasculitis.[5] PAN is sometimes associated with infection by the hepatitis B or hepatitis C virus.[6] The condition may be present in infants.[7]
PAN is a rare disease.[6] With treatment, five-year survival is 80%; without treatment, five-year survival is 13%. Death is often a consequence of kidney failure, myocardial infarction, or stroke.[8]
https://en.wikipedia.org/wiki/Polyarteritis_nodosa
https://en.wikipedia.org/wiki/Black_hairy_tongue
https://en.wikipedia.org/wiki/Clonal_hematopoiesis
https://en.wikipedia.org/wiki/List_of_contaminated_cell_lines
Tartrate-resistant acid phosphatase (TRAP or TRAPase), also called acid phosphatase 5, tartrate resistant (ACP5), is a glycosylated monomeric metalloprotein enzyme expressed in mammals.[3] It has a molecular weight of approximately 35kDa, a basic isoelectric point (7.6–9.5), and optimal activity in acidic conditions. TRAP is synthesized as latent proenzyme and activated by proteolytic cleavage and reduction.[4][5] It is differentiated from other mammalian acid phosphatases by its resistance to inhibition by tartrate and by its molecular weight.
The mechanism of phosphate ester hydrolysis by TRAP is through a nucleophilic attack mechanism,[6] whereby, catalysis occurs with the binding of a phosphate-substrate to the Fe2+ in the active site of TRAP. This is then followed by a nucleophilic attack by a hydroxide ligand on the bound phosphorus atom, resulting in cleavage of the phosphate ester bond and production of an alcohol. The exact identity and mechanism of the hydroxide ligand is unclear, but it is thought to be either a hydroxide that bridges the metal ions within the active site or a terminal hydroxide bound to Fe3+, with conflicting reports for both mechanisms.
https://en.wikipedia.org/wiki/Tartrate-resistant_acid_phosphatase
Hairy root culture, also called transformed root culture, is a type of plant tissue culture that is used to study plant metabolic processes or to produce valuable secondary metabolites or recombinant proteins, often with plant genetic engineering.[1]
A naturally occurring soil bacterium Agrobacterium rhizogenes that contains root-inducing plasmids (also called Ri plasmids) can infect plant roots and cause them to produce a food source for the bacterium, opines, and to grow abnormally.[2] The abnormal roots are particularly easy to culture in artificial media because hormones are not needed in contrast to adventitious roots,[2] and they are neoplastic, with indefinite growth. The neoplastic roots produced by A. rhizogenes infection have a high growth rate (compared to untransformed adventitious roots), as well as genetic and biochemical stability.
Currently the main constraint for commercial utilization of hairy root culture is the development and up-scaling of appropriate (bioreactors) vessels for the delicate and sensitive hairy roots.[2][3][4]
Some of the applied research on utilization of hairy root cultures has been and is conducted at VTT Technical Research Centre of Finland Ltd.[5][6] Other labs working on hairy roots are the phytotechnology lab of Amiens University and the Arkansas Biosciences Institute.
https://en.wikipedia.org/wiki/Hairy_root_culture
https://en.wikipedia.org/wiki/List_of_skin_conditions
The Hairy Ape is a 1922 expressionist play by American playwright Eugene O'Neill. It is about a beastly, unthinking laborer known as Yank, the protagonist of the play, as he searches for a sense of belonging in a world controlled by the rich. At first, Yank feels secure as he stokes the engines of an ocean liner, and is highly confident in his physical power over the ship's engines and his men.
However, when the rich daughter of an industrialist in the steel business refers to him as a "filthy beast", Yank undergoes a crisis of identity and so starts his mental and physical deterioration. He leaves the ship and wanders into Manhattan, only to find he does not belong anywhere—neither with the socialites on Fifth Avenue, nor with the labor organizers on the waterfront. In a fight for social belonging, Yank's mental state disintegrates into animalistic, and in the end, he is defeated by an ape in which Yank's character has been reflected. The Hairy Ape is a portrayal of the impact industrialization and social class has on the dynamic character Yank.
https://en.wikipedia.org/wiki/The_Hairy_Ape
List of human clusters of differentiation
https://en.wikipedia.org/wiki/List_of_human_clusters_of_differentiation
Acute necrotizing ulcerative gingivitis (ANUG) is a common, non-contagious infection of the gums with sudden onset. The main features are painful, bleeding gums, and ulceration of inter-dental papillae (the sections of gum between adjacent teeth). This disease, along with necrotizing (ulcerative) periodontitis (NP or NUP) is classified as a necrotizing periodontal disease, one of the seven general types of gum disease caused by inflammation of the gums (periodontitis).
The often severe gum pain that characterizes ANUG distinguishes it from the more common chronic periodontitis which is rarely painful. If ANUG is improperly treated or neglected, it may become chronic and/or recurrent. The causative organisms are mostly anaerobic bacteria, particularly Fusobacteriota and spirochete species.
Predisposing factors include poor oral hygiene, smoking, poor nutrition, psychological stress, and a weakened immune system. When the attachments of the teeth to the bone are involved, the term NUP is used. Treatment of ANUG is by removal of dead gum tissue and antibiotics (usually metronidazole) in the acute phase, and improving oral hygiene to prevent recurrence. Although the condition has a rapid onset and is debilitating, it usually resolves quickly and does no serious harm. The informal name trench mouth arose during World War I as many soldiers developed the disease, probably because of the poor conditions and extreme psychological stress.
https://en.wikipedia.org/wiki/Acute_necrotizing_ulcerative_gingivitis
Hairy skin
The hairy parts of the body such as the forearm or the leg have two groups of nerve endings: those that end along with the hair follicles, and also with the arborizations of unmyelinated axons which are referred to as free nerve endings because they are served by both myelinated and unmyelinated axons.[1]
https://en.wikipedia.org/wiki/Cutaneous_innervation#Hairy_skin
Glyphosate (IUPAC name: N-(phosphonomethyl)glycine) is a broad-spectrum systemic herbicide and crop desiccant. It is an organophosphorus compound, specifically a phosphonate, which acts by inhibiting the plant enzyme 5-enolpyruvylshikimate-3-phosphate synthase. It is used to kill weeds, especially annual broadleaf weeds and grasses that compete with crops. Its herbicidal effectiveness was discovered by Monsanto chemist John E. Franz in 1970. Monsanto brought it to market for agricultural use in 1974 under the trade name Roundup. Monsanto's last commercially relevant United States patent expired in 2000.
Farmers quickly adopted glyphosate for agricultural weed control, especially after Monsanto introduced glyphosate-resistant Roundup Ready crops, enabling farmers to kill weeds without killing their crops. In 2007, glyphosate was the most used herbicide in the United States' agricultural sector and the second-most used (after 2,4-D) in home and garden, government and industry, and commercial applications.[6] From the late 1970s to 2016, there was a 100-fold increase in the frequency and volume of application of glyphosate-based herbicides (GBHs) worldwide, with further increases expected in the future.
Glyphosate is absorbed through foliage, and minimally through roots, and transported to growing points. It inhibits a plant enzyme involved in the synthesis of three aromatic amino acids: tyrosine, tryptophan, and phenylalanine. It is therefore effective only on actively growing plants and is not effective as a pre-emergence herbicide. An increasing number of crops have been genetically engineered to be tolerant of glyphosate (e.g. Roundup Ready soybean, the first Roundup Ready crop, also created by Monsanto), which allows farmers to use glyphosate as a post-emergence herbicide against weeds.
While glyphosate and formulations such as Roundup have been approved by regulatory bodies worldwide, concerns about their effects on humans and the environment have persisted.[7][8] A number of regulatory and scholarly reviews have evaluated the relative toxicity of glyphosate as an herbicide. The WHO and FAO Joint committee on pesticide residues issued a report in 2016 stating the use of glyphosate formulations does not necessarily constitute a health risk, and giving an acceptable daily intake limit of 1 milligram per kilogram of body weight per day for chronic toxicity.[9]
The consensus among national pesticide regulatory agencies and scientific organizations is that labeled uses of glyphosate have demonstrated no evidence of human carcinogenicity.[10] The German Federal Institute for Risk Assessment toxicology review in 2013 found that with regard to positive correlations between exposure to glyphosate formulations and risk of various cancers, including non-Hodgkin lymphoma, "the available data is contradictory and far from being convincing".[11] A meta-analysis published in 2014 identified an increased risk of NHL in workers exposed to glyphosate formulations.[12] In March 2015, the World Health Organization's International Agency for Research on Cancer (IARC) classified glyphosate as "probably carcinogenic in humans" (category 2A) based on epidemiological studies, animal studies, and in vitro studies.[8][13][14][15] In contrast, the European Food Safety Authority concluded in November 2015 that "the substance is unlikely to be genotoxic (i.e. damaging to DNA) or to pose a carcinogenic threat to humans", later clarifying that while carcinogenic glyphosate-containing formulations may exist, studies "that look solely at the active substance glyphosate do not show this effect."[16][17] In 2017, the European Chemicals Agency (ECHA) classified glyphosate as causing serious eye damage and as toxic to aquatic life, but did not find evidence implicating it as a carcinogen, a mutagen, toxic to reproduction, nor toxic to specific organs.[18]
https://en.wikipedia.org/wiki/Glyphosate
A pyogenic granuloma or lobular capillary hemangioma[3] is a vascular tumor that occurs on both mucosa and skin, and appears as an overgrowth of tissue due to irritation, physical trauma, or hormonal factors.[4][5] It is often found to involve the gums, skin, or nasal septum, and has also been found far from the head, such as in the thigh.[6]
Pyogenic granulomas may be seen at any age, and are more common in females than males. In pregnant women, lesions may occur in the first trimester with an increasing incidence until the seventh month, and are often seen on the gums.[7]
https://en.wikipedia.org/wiki/Pyogenic_granuloma
Y linkage, also known as holandric inheritance (from Ancient Greek ὅλος hólos, "whole" + ἀνδρός andrós, "male"),[1] describes traits that are produced by genes located on the Y chromosome. It is a form of sex linkage.
Y linkage can be difficult to detect. This is partly because the Y chromosome is small and contains fewer genes than the autosomal chromosomes or the X chromosome. It is estimated to contain about 200 genes. Earlier, the human Y chromosome was thought to have little importance;.[2] Although the Y-chromosome is sex-determining in humans and some other species, not all genes that play a role in sex determination are Y-linked. The Y-chromosome, generally does not undergo genetic recombination and only small regions called pseudoautosomal regions exhibit recombination. The majority of the Y-chromosome genes that do not recombine are located in the "non-recombining region".[3]
For a trait to be considered Y linkage, it must exhibit these characteristics:
- occurs only in males
- appears in all sons of males who exhibit that trait
- is absent from daughters of trait carriers; instead the daughters that are phenotypically normal and do not have affected offspring.[4]
These requirements were established by the pioneer of Y linkage, Curt Stern. Stern detailed in his paper genes he suspected to be Y-linked.[4] His requirements at first made Y linkage hard to prove. In the 1950s using human pedigrees, many genes were incorrectly determined to be Y-linked.[5] Later research adopted more advanced techniques and more sophisticated statistical analysis.[6] Hairy ears are an example of a gene once thought to be Y-linked in humans; however, that hypothesis was discredited.[5] Due to advancements in DNA sequencing, Y linkage is getting easier to determine and prove. The Y-chromosome is almost entirely mapped, revealing many Y-linked traits.
Y linkage is similar to, but different from X linkage; although, both are forms of sex linkage. X linkage can be genetically linked and sex-linked, while Y linkage can only be genetically linked. This is because males' cells have only one copy of the Y-chromosome. X-chromosomes have two copies, one from each parent permitting recombination. The X chromosome contains more genes and is substantially larger.
Some ostensibly Y-linked traits have not been confirmed. One example is hearing impairment. Hearing impairment was tracked in one specific family and through seven generations all males were affected by this trait. However, this trait occurs rarely and has not been entirely resolved.[7]
Y-chromosome deletions are a frequent genetic cause of male infertility.
https://en.wikipedia.org/wiki/Y_linkage
A bioreactor refers to any manufactured device or system that supports a biologically active environment.[1] In one case, a bioreactor is a vessel in which a chemical process is carried out which involves organisms or biochemically active substances derived from such organisms. This process can either be aerobic or anaerobic. These bioreactors are commonly cylindrical, ranging in size from litres to cubic metres, and are often made of stainless steel.[citation needed] It may also refer to a device or system designed to grow cells or tissues in the context of cell culture.[2] These devices are being developed for use in tissue engineering or biochemical/bioprocess engineering.[citation needed]
On the basis of mode of operation, a bioreactor may be classified as batch, fed batch or continuous (e.g. a continuous stirred-tank reactor model). An example of a continuous bioreactor is the chemostat.[citation needed]
Organisms or biochemically active substances growing in bioreactors may be submerged in liquid medium or may be anchored to the surface of a solid medium. Submerged cultures may be suspended or immobilized. Suspension bioreactors may support a wider variety of organisms, since special attachment surfaces are not needed, and can operate at a much larger scale than immobilized cultures. However, in a continuously operated process the organisms will be removed from the reactor with the effluent. Immobilization is a general term describing a wide variety of methods for cell or particle attachment or entrapment.[3] It can be applied to basically all types of biocatalysis including enzymes, cellular organelles, animal and plant cells and organs.[4][5] Immobilization is useful for continuously operated processes, since the organisms will not be removed with the reactor effluent, but is limited in scale because the microbes are only present on the surfaces of the vessel.
Large scale immobilized cell bioreactors are:
- moving media, also known as moving bed biofilm reactor (MBBR)
- packed bed
- fibrous bed
- membrane
https://en.wikipedia.org/wiki/Bioreactor
Plant tissue culture is a collection of techniques used to maintain or grow plant cells, tissues, or organs under sterile conditions on a nutrient culture medium of known composition. It is widely used, to produce clones of a plant in a method known as micropropagation. Different techniques in plant tissue culture may offer certain advantages over traditional methods of propagation, including:
- The production of exact copies of plants that produce particularly good flowers, fruits, or other desirable traits.
- To quickly produce mature plants.
- The production of multiples of plants in the absence of seeds or necessary pollinators to produce seeds.
- The regeneration of whole plants from plant cells that have been genetically modified.
- The production of plants in sterile containers allows them to be moved with greatly reduced chances of transmitting diseases, pests, and pathogens.
- The production of plants from seeds that otherwise have very low chances of germinating and growing, i.e. orchids and Nepenthes.
- To clean particular plants of viral and other infections and to quickly multiply these plants as 'cleaned stock' for horticulture and agriculture.
- Reproduce recalcitrant plants required for land restoration
- Storage of genetic plant material to safeguard native plant species.
Plant tissue culture relies on the fact that many plant parts have the ability to regenerate into a whole plant (cells of those regenerative plant parts are called totipotent cells which can differentiate into various specialized cells). Single cells, plant cells without cell walls (protoplasts), pieces of leaves, stems or roots can often be used to generate a new plant on culture media given the required nutrients and plant hormones.
https://en.wikipedia.org/wiki/Plant_tissue_culture
Trametes hirsuta, commonly known as hairy bracket, is a fungal plant pathogen. It is found on dead wood of deciduous trees, especially beechwood. It is found all year round and persists due to its leathery nature.[1]
The cap is whitish gray, with short hairs, sometimes yellowish and tomentose at the edge, and with subtle zoning. The flesh is tough with a soft gray upper layer and a whitish lower layer, separated by a black plane.[2]
Similar species include T. pubescens, which is unzoned, buff in colour, and without layered flesh. T. versicolor is more distinctively zoned.[2]
https://en.wikipedia.org/wiki/Trametes_hirsuta
The Compensation scheme for radiation-linked diseases is a workers compensation scheme administered by the UK government. It was established in November 1982 by British Nuclear Fuels Limited and its trade unions[1] following legal actions brought against the company by nuclear industry workers in the late 1970s. At the time of its establishment, BNFL and its trade unions agreed that the causation of cancer by radiation was sufficiently well understood that "it should be possible to construct a scheme which would evaluate the probability that a diagnosed cancer may have been caused by radiation exposure at work." Initially the scheme only accepted claims in which a worker had died from a radiation-linked disease. In 1987 this was expanded to allow morbidity claims.[2] The list of participating member employers and trade unions has grown through the 1990s and 2000s.[3] As of July 2021, 1710 cases had been received and considerered since the Scheme began. 163 of those resulted in successful claims. Compensation payments exceeding £8.90 million have been paid to claimants.[4]
https://en.wikipedia.org/wiki/Compensation_scheme_for_radiation-linked_diseases
The following outline is provided as an overview of and topical guide to immunology:
Immunology is the study of all aspects of the immune system in all organisms.[1] It deals with the physiological functioning of the immune system in states of both health and disease; malfunctions of the immune system in immunological disorders (autoimmune diseases, hypersensitivities, immune deficiency, transplant rejection); the physical, chemical and physiological characteristics of the components of the immune system in vitro, in situ, and in vivo.
https://en.wikipedia.org/wiki/Outline_of_immunology
Epulis (Greek: ἐπουλίς; plural epulides) is any tumor like enlargement (i.e. lump) situated on the gingival or alveolar mucosa.[1][2] The word literally means "(growth) on the gingiva",[3][4] and describes only the location of the mass and has no further implications on the nature of the lesion.[5] There are three types: fibromatous, ossifying and acanthomatous.[medical citation needed] The related term parulis (commonly called a gumboil) refers to a mass of inflamed granulation tissue at the opening of a draining sinus on the alveolus over (or near to) the root of an infected tooth.[2] Another closely related term is gingival enlargement, which tends to be used where the enlargement is more generalized over the whole gingiva rather than a localized mass.
https://en.wikipedia.org/wiki/Epulis
| Hypertrichosis | |
|---|---|
| Other names | Werewolf syndrome |
 | |
| Petrus Gonsalvus, "’The Hairy Man’", as illustrated by Joris Hoefnagel in his Elementa Depicta | |
| Specialty | Dermatology |
Hypertrichosis is an abnormal amount of hair growth over the body.[1][2] The two distinct types of hypertrichosis are generalized hypertrichosis, which occurs over the entire body, and localized hypertrichosis, which is restricted to a certain area.[1] Hypertrichosis can be either congenital (present at birth) or acquired later in life.[3][4] The excess growth of hair occurs in areas of the skin with the exception of androgen-dependent hair of the pubic area, face, and axillary regions.[5]
Several circus sideshow performers in the 19th and early 20th centuries, such as Julia Pastrana, had hypertrichosis.[6] Many of them worked as freaks and were promoted as having distinct human and animal traits.
https://en.wikipedia.org/wiki/Hypertrichosis
Arthropods, including insects and spiders, make use of smooth adhesive pads as well as hairy pads for climbing and locomotion along non-horizontal surfaces.[1][2][3] Both types of pads in insects make use of liquid secretions and are considered 'wet'.[3] Dry adhesive mechanisms primarily rely on Van der Waals' forces and are also used by organisms other than insects.[4] The fluid provides capillary and viscous adhesion and appears to be present in all insect adhesive pads.[5] Little is known about the chemical properties of the adhesive fluids and the ultrastructure of the fluid-producing cells is currently not extensively studied.[4] Additionally, both hairy and smooth types of adhesion have evolved separately numerous times in insects.[3][6] Few comparative studies between the two types of adhesion mechanisms have been done and there is a lack of information regarding the forces that can be supported by these systems in insects.[3] Additionally, tree frogs and some mammals such as the arboreal possum and bats also make use of smooth adhesive pads.[1][2] The use of adhesive pads for locomotion across non-horizontal surfaces is a trait that evolved separately in different species, making it an example of convergent evolution.[7] The power of adhesion allows these organisms to be able to climb on almost any substance.[2]
The exact mechanisms of arthropod adhesion are still unknown for some species but this topic is of great importance to biologists, physicists and engineers.[2][3][7] These highly specialized structures are not restricted to one particular area of the leg. They may be located on different parts, such as claws, derivatives of the pretarsus, tarsal apex, tarsomeres or tibia.[6] From the scaling analysis, it has been suggested that animal lineages relying on the dry adhesion, such as lizards and spiders have a higher density of terminal contact elements compared to systems that use wet adhesive mechanisms such as insects.[6] Since these effects are based on fundamental physical principles and highly related to the shape of the structure, they are also the same for artificial surfaces with similar geometry.[6] Adhesion and friction forces per-unit-pad area were very similar in smooth and hairy systems when tested.[3] Strong adhesion may be beneficial in many situations but it also can create difficulties in locomotion.[3] Direction-dependence is an important and fundamental property of adhesive structures that are able to rapidly and controllably adhere during locomotion.[3] Researchers are unsure whether direction-dependence is achieved through changes in contact area or through a change in shear stress.[3] Friction and adhesion forces in most animal attachment organs are higher when they are pulled towards the body than when they push away from it.[3] This has been observed in geckos and spiders but also in the smooth adhesive pads of ants, bush-crickets and cockroaches.[3] Adhesive hairs of geckos are non-symmetrical and feature distally pointing setae and spatulae that are able to generate increased friction and adhesion when aligned with a proximal pull.[3] The adhesive hairs of some beetles behave similarly to those of geckos.[3] While directional-dependence is present in other animals, it has yet to be confirmed in insects with hairy adhesive pads.[3]
It has been observed that a surface micro-roughness asperity size of less than five micrometres can strongly reduce insect attachment and climbing ability and this adhesion reducing effect has been put to use in a variety of plant species that create wax crystals.[5]
Adhesive chemical secretions are also used for predation defence, mating, holding substrates, anchor eggs, building retreats, prey capture and self-grooming.[4]
https://en.wikipedia.org/wiki/Arthropod_adhesion
| Peripheral giant-cell granuloma | |
|---|---|
| Specialty | Dentistry, ENT surgery |
Peripheral giant-cell granuloma (PGCG) is an oral pathologic condition that appears in the mouth as an overgrowth of tissue due to irritation or trauma. Because of its overwhelming incidence on the gingiva, the condition is associated with two other diseases, pyogenic granuloma and peripheral ossifying fibroma. These three diseases are associated because they appear frequently on gingiva. Due to its similar microscopic appearance, peripheral giant-cell granuloma is considered to be the soft tissue equivalent of central giant-cell granuloma.
The appearance of peripheral giant-cell granuloma is also similar to pyogenic granuloma. The color ranges from red to bluish-purple, but is usually more blue in comparison to pyogenic granuloma. It can be sessile or pedunculated with the size usually being less than 2 cm.
The lesion has a 60% gender predilection to females. The prevalence of the peripheral giant-cell granuloma is highest around 50 - 60 years of age. It appears only on the gingiva or on an edentulous alveolar ridge. It is more often found in the mandible rather than the maxilla, in either anterior or posterior areas. The underlying alveolar bone can be destroyed, leaving a unique appearance referred to as "cupping resorption" or "saucerization".
https://en.wikipedia.org/wiki/Peripheral_giant-cell_granuloma
| |||||||
| |||||||
| |||||||
| |||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
TGF beta-inducible nuclear protein 1 is a protein that in humans is encoded by the NSA2 gene.[5][6][7]
https://en.wikipedia.org/wiki/TINP1
Rhizomucor pusillus is a species of Rhizomucor. It can cause disease in humans.[1][2] R. pusillus is a grey mycelium fungi most commonly found in compost piles. Yellow-brown spores grow on a stalk to reproduce more fungal cells.[clarification needed]
https://en.wikipedia.org/wiki/Rhizomucor_pusillus
https://en.wikipedia.org/wiki/List_of_common_misconceptions
https://en.wikipedia.org/wiki/Brain%E2%80%93computer_interface#Cell-culture_BCIs
https://en.wikipedia.org/wiki/Hemipenthes_morio
https://en.wikipedia.org/wiki/Stafne_defect
https://en.wikipedia.org/wiki/Microdontia
https://en.wikipedia.org/wiki/Tunga_penetrans
https://en.wikipedia.org/wiki/Condylar_resorption
https://en.wikipedia.org/wiki/Idiopathic_osteosclerosis
https://en.wikipedia.org/wiki/Flagellum
https://en.wikipedia.org/wiki/Neonatal_teeth
https://en.wikipedia.org/wiki/Pneumoparotitis
https://en.wikipedia.org/wiki/Cat
https://en.wikipedia.org/wiki/Segmentation_(biology)
https://en.wikipedia.org/wiki/Transfer_DNA
https://en.wikipedia.org/wiki/List_of_MeSH_codes_(C15)
https://en.wikipedia.org/wiki/Darier%27s_disease
https://en.wikipedia.org/wiki/List_of_animals_that_can_get_SARS-CoV-2
https://en.wikipedia.org/wiki/Guar
https://en.wikipedia.org/wiki/Phenoxymethylpenicillin
https://en.wikipedia.org/wiki/Pulp_stone