Amidophosphoribosyltransferase (ATase), also known as glutamine phosphoribosylpyrophosphate amidotransferase (GPAT), is an enzyme responsible for catalyzing the conversion of 5-phosphoribosyl-1-pyrophosphate (PRPP) into 5-phosphoribosyl-1-amine (PRA), using the amine group from a glutamine side-chain. This is the committing step in de novo purine synthesis. In humans it is encoded by the PPAT (phosphoribosyl pyrophosphate amidotransferase) gene.[5][6] ATase is a member of the purine/pyrimidine phosphoribosyltransferase family.
https://en.wikipedia.org/wiki/Amidophosphoribosyltransferase
In enzymology, a glutamate N-acetyltransferase (EC 2.3.1.35) is an enzyme that catalyzes the chemical reaction
- N2-acetyl-L-ornithine + L-glutamate L-ornithine + N-acetyl-L-glutamate
Thus, the two substrates of this enzyme are N2-acetyl-L-ornithine and L-glutamate, whereas its two productsare L-ornithine and N-acetyl-L-glutamate.
This enzyme belongs to the family of transferases, specifically those acyltransferases transferring groups other than aminoacyl groups. The systematic name of this enzyme class is N2-acetyl-L-ornithine:L-glutamate N-acetyltransferase. Other names in common use include ornithine transacetylase, alpha-N-acetyl-L-ornithine:L-glutamate N-acetyltransferase, acetylglutamate synthetase, acetylglutamate-acetylornithine transacetylase, acetylglutamic synthetase, acetylglutamic-acetylornithine transacetylase, acetylornithinase, acetylornithine glutamate acetyltransferase, glutamate acetyltransferase, N-acetyl-L-glutamate synthetase, N-acetylglutamate synthase, N-acetylglutamate synthetase, ornithine acetyltransferase, and 2-N-acetyl-L-ornithine:L-glutamate N-acetyltransferase. This enzyme participates in urea cycle and metabolism of amino groups.
https://en.wikipedia.org/wiki/Glutamate_N-acetyltransferase
Brucella anthropi is a bacterium.[3] The type strain is strain CIP 82.115 (= CIP 14970 = NCTC 12168 = LMG 3331). O. anthropi strains are rod-shaped, aerobic, gram-negative, non-pigmented and motile by means of peritrichous flagella.[4][5][6] They are emerging as major opportunistic pathogens.[7]
https://en.wikipedia.org/wiki/Brucella_anthropi
Vacuolar-type ATPase (V-ATPase) is a highly conserved evolutionarily ancient enzyme with remarkably diverse functions in eukaryotic organisms.[1] V-ATPases acidify a wide array of intracellular organelles and pump protons across the plasma membranes of numerous cell types. V-ATPases couple the energy of ATP hydrolysis to proton transport across intracellular and plasma membranes of eukaryotic cells. It is generally seen as the polar opposite of ATP synthase because ATP synthase is a proton channel that uses the energy from a proton gradient to produce ATP. V-ATPase however, is a proton pump that uses the energy from ATP hydrolysis to produce a proton gradient.
The Archaea-type ATPase (A-ATPase) is a related group of ATPases found in Archaea that often work as an ATP synthase. It forms a clade V/A-ATPase with V-ATPase. Most members of either group shuttle protons (H+
), but a few members have evolved to use sodium ions (Na+
) instead.
| V-ATPase | |
|---|---|
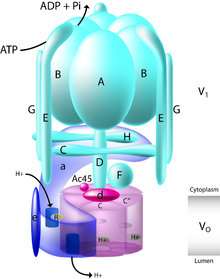 V-ATPase schematic | |
| Identifiers | |
| Symbol | V-ATPase |
| TCDB | 3.A.2 |
| OPM superfamily | 5 |
| OPM protein | 2bl2 |
| Membranome | 226 |
The overall reaction catalyzed by ATase is the following:
- PRPP + glutamine → PRA + glutamate + PPi
Within the enzyme, the reaction is broken down into two half-reactions that occur at different active sites:
- glutamine → NH
3 + glutamate - PRPP + NH
3 → PRA + PPi
The first part of the mechanism occurs in the active site of the glutaminase domain and releases an ammonia group from glutamine by hydrolysis. The ammonia released by the first reaction is then transferred to the active site of the phosphoribosyltransferase domain via a 20 Å channel, where it then binds to PRPP to form PRA.
https://en.wikipedia.org/wiki/V-ATPase
Gastric hydrogen potassium ATPase, also known as H+/K+ ATPase, is an enzyme which functions to acidify the stomach.[1] It is a member of the P-type ATPases, also known as E1-E2 ATPases due to its two states.[2]
https://en.wikipedia.org/wiki/Hydrogen_potassium_ATPase
In the field of enzymology, a proton ATPase is an enzyme that catalyzes the following chemical reaction:
- ATP + H
2O + H+
in ADP + phosphate + H+
out
The 3 substrates of this enzyme are ATP, H
2O, and H+
, whereas its 3 products are ADP, phosphate, and H+
.
Proton ATPases are divided into three groups[1] as outlined below:
https://en.wikipedia.org/wiki/Proton_ATPase
above.

No comments:
Post a Comment