chole palm black blud and the spindel scell scaroma sarcoma.
08-28-2021-1057 - Crustaceans daphne Daphniidae
Search Results
Page Navigation
| Previous | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | Next |
Phoronids (scientific name Phoronida, sometimes called horseshoe worms) are a small phylum of marine animals that filter-feed with a lophophore (a "crown" of tentacles), and build upright tubes of chitin to support and protect their soft bodies. They live in most of the oceans and seas, including the Arctic Ocean but excluding the Antarctic Ocean, and between the intertidal zone and about 400 meters down. Most adult phoronids are 2 cm long and about 1.5 mm wide, although the largest are 50 cm long.
The name of the group comes from its type genus: Phoronis.[2][3]
| Phoronids | |
|---|---|
 |
https://en.wikipedia.org/wiki/Phoronid
The Spiralia are a morphologically diverse clade of protostome animals, including within their number the molluscs, annelids, platyhelminths and other taxa.[1] The term Spiralia is applied to those phyla that exhibit canonical spiral cleavage, a pattern of early development found in most (but not all) members of the Lophotrochozoa.[2]
Distribution of spiralian development across phylogeny[edit]
Members of the molluscs, annelids, platyhelminths and nemerteans have all been shown to exhibit spiral cleavage in its classical form. Other spiralian phyla (rotifers, brachiopods, phoronids, gastrotrichs, and bryozoans) are also said to display a derived form of spiral cleavage in at least a portion of their constituent species, although evidence for this is sparse.[3]
| Spiralia | |
|---|---|
 |
The gastrotrichs (phylum Gastrotricha), commonly referred to as hairybellies or hairybacks, are a group of microscopic (0.06-3.0 mm), worm-like, pseudocoelomate animals, and are widely distributed and abundant in freshwater and marine environments. They are mostly benthic and live within the periphyton, the layer of tiny organisms and detritus that is found on the seabed and the beds of other water bodies. The majority live on and between particles of sediment or on other submerged surfaces, but a few species are terrestrial and live on land in the film of water surrounding grains of soil. Gastrotrichs are divided into two orders, the Macrodasyida which are marine (except for two species), and the Chaetonotida, some of which are marine and some freshwater. Nearly 800 species of gastrotrich have been described.
Gastrotrichs have a simple body plan with a head region, with a brain and sensory organs, and a trunk with a simple gut and the reproductive organs. They have adhesive glands with which they can anchor themselves to the substrate and cilia with which they move around. They feed on detritus, sucking up organic particles with their muscular pharynx. They are hermaphrodites, the marine species producing eggs which develop directly into miniature adults. The freshwater species are parthenogenetic, producing unfertilised eggs, and at least one species is viviparous. Gastrotrichs mature with great rapidity and have lifespans of only a few days.
| Gastrotrich | |
|---|---|
 | |
| Darkfield photograph of a gastrotrich |
Gastrotrichs are cosmopolitan in distribution. They inhabit the interstitial spaces between particles in marine and freshwater environments, the surfaces of aquatic plants and other submerged objects and the surface film of water surrounding soil particles on land.[4] They are also found in stagnant pools and anaerobic mud, where they thrive even in the presence of hydrogen sulfide. When pools dry up they can survive periods of desiccation as eggs, and some species are capable of forming cysts in harsh conditions.[9] In marine sediments they have been known to reach 364 individuals per 10 cm2 (1.6 sq in) making them the third most common invertebrate in the sediment after nematodes and harpacticoid copepods. In freshwater they may reach a density of 158 individuals per 10 cm2 (1.6 sq in) and are the fifth most abundant group of invertebrates in the sediment.[4]
In marine and freshwater environments, gastrotrichs form part of the benthic community. They are detritivores and are microphagous, sucking dead or living organic material, diatoms, bacteria and small protozoa into their mouths by the muscular action of the pharynx. They are themselves eaten by turbellarians and other small macrofauna.[4]
Gastrotrichs vary in size from about 0.06 to 3 mm (0.002 to 0.118 in) in body length.[4] They are bilaterally symmetrical, with a transparent strap-shaped or bowling pin-shaped body, arched dorsally and flattened ventrally. The anterior end is not clearly defined as a head but contains the sense organs, brain and pharynx. Cilia are found around the mouth and on the ventral surface of the head and body. The trunk contains the gut and the reproductive organs. At the posterior end of the body are two projections with cement glands that serve in adhesion. This is a double-gland system where one gland secretes the glue and another secretes a de-adhesive agent to sever the connection. In the Macrodasyida, there are additional adhesive glands at the anterior end and on the sides of the body.[6]
The body wall consists of a cuticle, an epidermis and longitudinal and circular bands of muscle fibres. In some primitive species, each epidermal cell has a single cilium, a feature shared only by the gnathostomulans. The whole ventral surface of the animal may be ciliated or the cilia may be arranged in rows, patches or transverse bands. The cuticle is locally thickened in some gastrotrichs and forms scales, hooks and spines. There is no coelom (body cavity) and the interior of the animal is filled with poorly differentiated connective tissue. In the macrodasyidans, Y-shaped cells, each containing a vacuole, surround the gut and may function as a hydrostatic skeleton.[6]
The mouth is at the anterior end and opens into an elongated muscular pharynx with a triangular or Y-shaped lumen, lined by myoepithelial cells. The pharynx opens into a cylindrical intestine, which is lined with glandular and digestive cells. The anus is located on the ventral surface close to the posterior of the body. In some species, there are pores in the pharynx opening to the ventral surface; these contain valves and may allow egestion of any excess water swallowed while feeding.[6]
In the chaetonotidans, the excretory system consists of a single pair of protonephridia, which open through separate pores on the lateral underside of the animal, usually in the midsection of the body. In the macrodasyidans, there are several pairs of these opening along the side of the body. Nitrogenous waste is probably excreted through the body wall, as part of respiration, and the protonephridia are believed to function mainly in osmoregulation.[6] Unusually, the protonephridia do not take the form of flame cells, but, instead, the excretory cells consist of a skirt surrounding a series of cytoplasmic rods that in turn enclose a central flagellum. These cells, termed cyrtocytes, connect to a single outlet cell which passes the excreted material into the protonephridial duct.[8]
As is typical for such small animals, there are no respiratory or circulatory organs. The nervous system is relatively simple. The brain consists of two ganglia, one on either side of the pharynx, connected by a commissure. From these lead a pair of nerve cords which run along either side of the body beside the longitudinal muscle bands. The primary sensory organs are the bristles and ciliated tufts of the body surface which function as mechanoreceptors. There are also ciliated pits on the head, simple ciliary photoreceptors and fleshy appendages which act as chemoreceptors.[6]
The name "gastrotrich" comes from the Greek γαστήρ gaster, meaning "stomach", and θρίξ thrix, meaning "hair".[2] The name was coined by the Russian zoologist Élie Metchnikoff in 1865.[1] The common name "hairyback" apparently arises from a mistranslation of "gastrotrich".[3]
The relationship of gastrotrichs to other phyla is unclear. Morphology suggests that they are close to the Gnathostomulida, the Rotifera, or the Nematoda. On the other hand, genetic studies place them as close relatives of the Platyhelminthes, the Ecdysozoa or the Lophotrochozoa.[4] As of 2011, around 790 species have been described.[5] The phylum contains a single class, divided into two orders: the Macrodasyida and the Chaetonotida.[6] Edward Ruppert et al. report that the Macrodasyida are wholly marine,[6] but two rare and poorly known species, Marinellina flagellata and Redudasys fornerise, are known from fresh water.[7] The Chaetonotida comprises both marine and freshwater species.[6]
The gastrotrichs (phylum Gastrotricha), commonly referred to as hairybellies or hairybacks, are a group of microscopic (0.06-3.0 mm), worm-like, pseudocoelomate animals, and are widely distributed and abundant in freshwater and marine environments. They are mostly benthic and live within the periphyton, the layer of tiny organisms and detritus that is found on the seabed and the beds of other water bodies. The majority live on and between particles of sediment or on other submerged surfaces, but a few species are terrestrial and live on land in the film of water surrounding grains of soil. Gastrotrichs are divided into two orders, the Macrodasyidawhich are marine (except for two species), and the Chaetonotida, some of which are marine and some freshwater. Nearly 800 species of gastrotrich have been described.
Gastrotrichs have a simple body plan with a head region, with a brain and sensory organs, and a trunk with a simple gut and the reproductive organs. They have adhesive glands with which they can anchor themselves to the substrate and cilia with which they move around. They feed on detritus, sucking up organic particles with their muscular pharynx. They are hermaphrodites, the marine species producing eggs which develop directly into miniature adults. The freshwater species are parthenogenetic, producing unfertilised eggs, and at least one species is viviparous. Gastrotrichs mature with great rapidity and have lifespans of only a few days.
Gastrotrich reproduction and reproductive behaviour has been little studied. That of macrodasiyds probably most represents that of the ancestral lineage and these more primitive gastrotrichs are simultaneous hermaphrodites, possessing both male and female sex organs. There is generally a single pair of gonads, the anterior portion of which contains sperm-producing cells and the posterior portion producing ova. The sperm is sometimes packaged in spermatophores and is released through male gonopores that open, often temporarily, on the underside of the animal, roughly two-thirds of the way along the body. A copulatory organ on the tail collects the sperm and transfers it to the partner's seminal receptacle through the female gonopore. Details of the process and the behaviour involved vary with the species, and there is a range of different accessory reproductive organs. During copulation, the "male" individual uses his copulatory organ to transfer sperm to his partner's gonopore and fertilisation is internal. The fertilised eggs are released by rupture of the body wall which afterwards repairs itself. As is the case in most protostomes, development of the embryo is determinate, with each cell destined to become a specific part of the animal's body.[6] At least one species of gastrotrich, Urodasys viviparus, is viviparous.[10]
Many species of chaetotonid gastrotrichs reproduce entirely by parthenogenesis. In these species, the male portions of the reproductive system are degenerate and non-functional, or, in many cases, entirely absent. Though the eggs have a diameter of less than 50 µm, they are still very large in comparison with the animals' size. Some species are capable of laying eggs that remain dormant during times of desiccation or low temperatures; these species, however, are also able to produce regular eggs, which hatch in one to four days, when environmental conditions are more favourable. The eggs of all gastrotrichs undergo direct development and hatch into miniature versions of the adult. The young typically reach sexual maturity in about three days. In the laboratory, Lepidodermella squamatum has lived for up to forty days, producing four or five eggs during the first ten days of life.[6]
Gastrotrichs demonstrate eutely, each species having an invariant genetically fixed number of cells as adults. Cell division ceases at the end of embryonic development and further growth is solely due to cell enlargement.[6]
https://en.wikipedia.org/wiki/Gastrotrich
Gastrotrichs are cosmopolitan in distribution. They inhabit the interstitial spaces between particles in marine and freshwater environments, the surfaces of aquatic plants and other submerged objects and the surface film of water surrounding soil particles on land.[4] They are also found in stagnant pools and anaerobic mud, where they thrive even in the presence of hydrogen sulfide. When pools dry up they can survive periods of desiccation as eggs, and some species are capable of forming cysts in harsh conditions.[9] In marine sediments they have been known to reach 364 individuals per 10 cm2 (1.6 sq in) making them the third most common invertebrate in the sediment after nematodes and harpacticoid copepods. In freshwater they may reach a density of 158 individuals per 10 cm2 (1.6 sq in) and are the fifth most abundant group of invertebrates in the sediment.[4]
https://en.wikipedia.org/wiki/Gastrotrich#Distribution_and_habitat
The rotifers (/ˈroʊtɪfərz/, from Latin rota "wheel" and -fer "bearing"), commonly called wheel animals or wheel animalcules,[1] make up a phylum (Rotifera /roʊˈtɪfərə/) of microscopic and near-microscopic pseudocoelomate animals.
They were first described by Rev. John Harris in 1696, and other forms were described by Antonie van Leeuwenhoek in 1703.[2] Most rotifers are around 0.1–0.5 mm long (although their size can range from 50 μmto over 2 mm),[1] and are common in freshwater environments throughout the world with a few saltwaterspecies.
Some rotifers are free swimming and truly planktonic, others move by inchworming along a substrate, and some are sessile, living inside tubes or gelatinous holdfasts that are attached to a substrate. About 25 species are colonial (e.g., Sinantherina semibullata), either sessile or planktonic. Rotifers are an important part of the freshwater zooplankton, being a major foodsource and with many species also contributing to the decomposition of soil organic matter.[3] Most species of the rotifers are cosmopolitan, but there are also some endemic species, like Cephalodella vittata to Lake Baikal.[4] Recent barcoding evidence, however, suggests that some 'cosmopolitan' species, such as Brachionus plicatilis, B. calyciflorus, Lecane bulla, among others, are actually species complexes.[5][6] In some recent treatments, rotifers are placed with acanthocephalans in a larger clade called Syndermata.
In June 2021, biologists reported the restoration of bdelloid rotifers after being frozen for 24,000 years in the Siberian permafrost.[7]
| Rotifera | |
|---|---|
 | |
| Rotifera | |
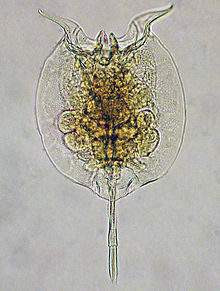 | |
| Pulchritia dorsicornuta |
https://en.wikipedia.org/wiki/Rotifer
Dicyemida, also known as Rhombozoa, is a phylum of tiny parasites that live in the renal appendages of cephalopods.
| Dicyemida | |
|---|---|
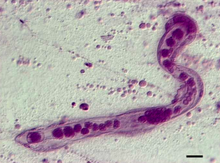 | |
| photomicrograph of Dicyema japonicum |
The Chaetognatha /kiːˈtɒɡnəθə/ or chaetognaths /ˈkiːtɒɡnæθs/ (meaning bristle-jaws) are a phylum of predatory marine worms that are a major component of plankton worldwide. Commonly known as arrow worms, about 20% of the known Chaetognatha species are benthic, and can attach to algae and rocks. They are found in all marine waters, from surface tropical waters and shallow tide pools to the deep sea and polar regions. Most chaetognaths are transparent and are torpedo shaped, but some deep-sea species are orange. They range in size from 2 to 120 millimetres (0.1 to 4.7 in).
There are more than 120 modern species assigned to over 20 genera.[3] Despite the limited diversity of species, the number of individuals is large.[4]
Arrow worms are usually considered a type of protostome that do not belong to either Ecdysozoa or Lophotrochozoa.
| Arrow worms | |
|---|---|
 | |
| Spadella cephaloptera |
| Loricifera | |
|---|---|
 | |
| Pliciloricus enigmaticus |
| Nematomorpha | |
|---|---|
 | |
| Paragordius tricuspidatus |
| Placozoa | |
|---|---|
 | |
| Trichoplax adhaerens |
Harpacticoida is an order of copepods, in the subphylum Crustacea. This order comprises 463 genera and about 3,000 species; its members are benthic copepods found throughout the world in the marine environment (most families) and in fresh water (essentially the Ameiridae, Parastenocarididae and the Canthocamptidae). A few of them are planktonic or live in association with other organisms. Harpacticoida represents the second-largest meiofaunal group in marine sediments, after nematodes. In Arctic and Antarctic seas, Harpacticoida are common inhabitants of sea ice. The name Harpacticoida comes from the Greek noun harpacticon (rapacious predator) and the suffix -oid (akin to) and means reminiscent of a predator .
Harpacticoids are distinguished from other copepods by the presence of only a very short pair of first antennae. The second pair of antennae are biramous, and the major joint within the body is located between the fourth and fifth body segments. They typically have a wide abdomen, and often have a somewhat worm-like body.[1]
| Harpacticoida | |
|---|---|
 | |
| Harpacticoid copepod |
https://en.wikipedia.org/wiki/Harpacticoida
Macrodasyida is an order of gastrotrichs.[1] Members of this order are somewhat worm-like in form, and not more than 1 to 1.5 mm in length.
Macrodasyids are almost in entirely marine and live in the sediment in marine or brackish water, but two species have been discovered in freshwater.[2] They can be distinguished from other gastrotrichs by the presence of two pores on either side of the pharynx, that allow excess water to be expelled during feeding. The body is dorsally flattened and there are tubular adhesive glands at both ends and on the lateral surfaces. These animals are detritivores and are hermaphrodites.[3]
| Macrodasyida | |
|---|---|
 | |
| Thaumastoderma ramuliferum (ventral view) |
https://en.wikipedia.org/wiki/Macrodasyida
The benthic zone is the ecological region at the lowest level of a body of water such as an ocean, lake, or stream, including the sediment surface and some sub-surface layers. Organisms living in this zone are called benthos and include microorganisms(e.g., bacteria and fungi)[1][2] as well as larger invertebrates, such as crustaceans and polychaetes.[3] Organisms here generally live in close relationship with the substrate and many are permanently attached to the bottom. The benthic boundary layer, which includes the bottom layer of water and the uppermost layer of sediment directly influenced by the overlying water, is an integral part of the benthic zone, as it greatly influences the biological activity that takes place there. Examples of contact soil layers include sand bottoms, rocky outcrops, coral, and bay mud.
| Marine habitats |
|---|
 Photomicrograph of typical benthic animals, including (from top to bottom) amphipods, a polychaete worm, a snail, and a chironomousmidge larva |
https://en.wikipedia.org/wiki/Benthic_zone
Amphipoda is an order of malacostracan crustaceans with no carapace and generally with laterally compressed bodies. Amphipods range in size from 1 to 340 millimetres (0.039 to 13 in) and are mostly detritivores or scavengers. There are more than 9,900 amphipod species so far described. They are mostly marine animals, but are found in almost all aquatic environments. Some 1,900 species live in fresh water, and the order also includes terrestrial animals and sandhoppers such as Talitrus saltator.
| Amphipoda | |
|---|---|
 | |
| Gammarus roeselii |
https://en.wikipedia.org/wiki/Amphipoda
Beggiatoa is a genus of Gammaproteobacteria belonging the order Thiotrichales, in the Proteobacteria phylum. This genus was one of the first bacteria discovered by Russian botanist Sergei Winogradsky. During his research in Anton de Bary’s laboratory of botany in 1887, he found that Beggiatoa oxidized hydrogen sulfide (H2S) as energy source, forming intracellular sulfur droplets, oxygen is the terminal electron acceptor and CO2 is used as carbon source. Winogradsky named it in honor of the Italian doctor and botanist Francesco Secondo Beggiato. Winogradsky referred to this form of metabolism as "inorgoxidation" (oxidation of inorganic compounds), today called chemolithotrophy. These organisms live in sulfur-rich environments such as soil, both marine and freshwater, in the deep sea hydrothermal vents and in polluted marine environments. The finding represented the first discovery of lithotrophy.[3][4] Two species of Beggiatoa have been formally described: the type species Beggiatoa alba and Beggiatoa leptomitoformis, the latter of which was only published in 2017.[2][5] This colorless and filamentous bacterium, sometimes in association with other sulfur bacteria (for example the genus Thiothrix), can be arranged in biofilm visible at naked eye formed by very long white filamentous mate, the white color is due to the stored sulfur. Species of Beggiatoa have cells up to 200 µ in diameter and they are one of the largest prokaryotes on Earth.[6]
| Beggiatoa | |
|---|---|
 | |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | |
| Family: | |
| Genus: | Beggiatoa Trevisan 1842[1] |
| Species | |
Beggiatoa alba | |
https://en.wikipedia.org/wiki/Beggiatoa
Vesicomyidae is a family of saltwater clams, marine bivalve molluscs in the superfamily Glossoidea.
| Vesicomyidae | |
|---|---|
 | |
| Vesicomya galatheae |
https://en.wikipedia.org/wiki/Vesicomyidae
A cold seep (sometimes called a cold vent) is an area of the ocean floor where hydrogen sulfide, methane and other hydrocarbon-rich fluid seepage occurs, often in the form of a brine pool. Cold does not mean that the temperature of the seepage is lower than that of the surrounding sea water. On the contrary, its temperature is often slightly higher.[1] The "cold" is relative to the very warm (at least 60 °C or 140 °F) conditions of a hydrothermal vent. Cold seeps constitute a biome supporting several endemic species.
Cold seeps develop unique topography over time, where reactions between methane and seawater create carbonate rock formations and reefs. These reactions may also be dependent on bacterial activity. Ikaite, a hydrous calcium carbonate, can be associated with oxidizing methane at cold seeps.
| Marine habitats |
|---|
 Tube worms are among the dominant species in one of four cold seep community types in the Gulf of Mexico. |
https://en.wikipedia.org/wiki/Cold_seep
Marine sediment, or ocean sediment, or seafloor sediment, are deposits of insoluble particles that have accumulated on the seafloor. These particles have their origins in soil and rocks and have been transported from the land to the sea, mainly by rivers but also by dust carried by wind and by the flow of glaciers into the sea. Additional deposits come from marine organisms and chemical precipitation in seawater, as well as from underwater volcanoes and meteorite debris.
Except within a few kilometres of a mid-ocean ridge, where the volcanic rock is still relatively young, most parts of the seafloor are covered in sediment. This material comes from several different sources and is highly variable in composition. Seafloor sediment can range in thickness from a few millimetres to several tens of kilometres. Near the surface seafloor sediment remains unconsolidated, but at depths of hundreds to thousands of metres the sediment becomes lithified(turned to rock).
Rates of sediment accumulation are relatively slow throughout most of the ocean, in many cases taking thousands of years for any significant deposits to form. Sediment transported from the land accumulates the fastest, on the order of one metre or more per thousand years for coarser particles. However, sedimentation rates near the mouths of large rivers with high discharge can be orders of magnitude higher. Biogenous oozes accumulate at a rate of about one centimetre per thousand years, while small clay particles are deposited in the deep ocean at around one millimetre per thousand years.
Sediments from the land are deposited on the continental margins by surface runoff, river discharge, and other processes. Turbidity currents can transport this sediment down the continental slope to the deep ocean floor. The deep ocean floor undergoes its own process of spreading out from the mid-ocean ridge, and then slowly subductsaccumulated sediment on the deep floor into the molten interior of the earth. In turn, molten material from the interior returns to the surface of the earth in the form of lava flowsand emissions from deep sea hydrothermal vents, ensuring the process continues indefinitely. The sediments provide habitat for a multitude of marine life, particularly of marine microorganisms. Their fossilized remains contain information about past climates, plate tectonics, ocean circulation patterns, and the timing of major extinctions.[1]
Within each colored area, the type of material shown is what dominates, although other materials are also likely to be present.
For further information about this diagram see below ↓
Except within a few kilometres of a mid-ocean ridge, where the volcanic rock is still relatively young, most parts of the seafloor are covered in sediments. This material comes from several different sources and is highly variable in composition, depending on proximity to a continent, water depth, ocean currents, biological activity, and climate. Seafloor sediments (and sedimentary rocks) can range in thickness from a few millimetres to several tens of kilometres. Near the surface, the sea-floor sediments remain unconsolidated, but at depths of hundreds to thousands of metres (depending on the type of sediment and other factors) the sediment becomes lithified.[2]
The various sources of seafloor sediment can be summarized as follows: [2]
- Terrigenous sediment is derived from continental sources transported by rivers, wind, ocean currents, and glaciers. It is dominated by quartz, feldspar, clay minerals, iron oxides, and terrestrial organic matter.
- Pelagic carbonate sediment is derived from organisms (e.g., foraminifera) living in the ocean water (at various depths, but mostly near surface) that make their shells (a.k.a. tests) out of carbonate minerals such as calcite.
- Pelagic silica sediment is derived from marine organisms (e.g., diatoms and radiolaria) that make their tests out of silica (microcrystalline quartz).
- Volcanic ash and other volcanic materials are derived from both terrestrial and submarine eruptions.
- Iron and manganese nodules form as direct precipitates from ocean-bottom water.
https://en.wikipedia.org/wiki/Marine_sediment
Aeolian processes, also spelled eolian, pertain to wind activity in the study of geology and weather and specifically to the wind's ability to shape the surface of the Earth (or other planets). Winds may erode, transport, and deposit materials and are effective agents in regions with sparse vegetation, a lack of soil moisture and a large supply of unconsolidated sediments. Although water is a much more powerful eroding force than wind, aeolian processes are important in arid environments such as deserts.[1]
The term is derived from the name of the Greek god Aeolus, the keeper of the winds.[2]
Particles are transported by winds through suspension, saltation (skipping or bouncing) and creeping (rolling or sliding) along the ground.
Small particles may be held in the atmosphere in suspension. Upward currents of air support the weight of suspended particles and hold them indefinitely in the surrounding air. Typical winds near Earth's surface suspend particles less than 0.2 millimeters in diameter and scatter them aloft as dust or haze.
Saltation is downwind movement of particles in a series of jumps or skips. Saltation normally lifts sand-size particles no more than one centimeter above the ground and proceeds at one-half to one-third the speed of the wind. A saltating grain may hit other grains that jump up to continue the saltation. The grain may also hit larger grains that are too heavy to hop, but that slowly creep forward as they are pushed by saltating grains. Surface creep accounts for as much as 25 percent of grain movement in a desert.
Aeolian turbidity currents are better known as dust storms. Air over deserts is cooled significantly when rain passes through it. This cooler and denser air sinks toward the desert surface. When it reaches the ground, the air is deflected forward and sweeps up surface debris in its turbulence as a dust storm.
Crops, people, villages, and possibly even climates are affected by dust storms. Some dust storms are intercontinental, a few may circle the globe, and occasionally they may engulf entire planets. When the Mariner 9 spacecraft entered its orbit around Mars in 1971, a dust storm lasting one month covered the entire planet, thus delaying the task of photo-mapping the planet's surface.[5]
Most of the dust carried by dust storms is in the form of silt-size particles. Deposits of this windblown silt are known as loess. The thickest known deposit of loess, 335 meters, is on the Loess Plateau in China. This very same Asian dust is blown for thousands of miles, forming deep beds in places as far away as Hawaii.[6] In Europe and in the Americas, accumulations of loess are generally from 20 to 30 meters thick. The soils developed on loess are generally highly productive for agriculture.
Aeolian transport from deserts plays an important role in ecosystems globally, e.g. by transport of minerals from the Sahara to the Amazon basin.[7] Saharan dust is also responsible for forming red clay soils in southern Europe.[8] Aeolian processes are affected by human activity, such as the use of 4x4 vehicles.[9]
Small whirlwinds, called dust devils, are common in arid lands and are thought to be related to very intense local heating of the air that results in instabilities of the air mass. Dust devils may be as much as one kilometer high.
https://en.wikipedia.org/wiki/Aeolian_processes#Transport
A dust storm, also called a sandstorm, is a meteorological phenomenon common in arid and semi-arid regions. Dust storms arise when a gust front or other strong wind blows loose sand and dirt from a dry surface. Fine particles are transported by saltation and suspension, a process that moves soil from one place and deposits it in another.
Drylands around North Africa and the Arabian peninsula are the main terrestrial sources of airborne dust. It has been argued that[1][unreliable source?] poor management of Earth's drylands, such as neglecting the fallow system, are increasing the size and frequency of dust storms from desert margins and changing both the local and global climate, and also impacting local economies.[2]
The term sandstorm is used most often in the context of desert dust storms, especially in the Sahara Desert, or places where sand is a more prevalent soil type than dirt or rock, when, in addition to fine particles obscuring visibility, a considerable amount of larger sand particles are blown closer to the surface. The term dust storm is more likely to be used when finer particles are blown long distances, especially when the dust storm affects urban areas.
Dust storms are not limited to Earth and have been known to form on other planets such as Mars.[18] These dust storms can extend over larger areas than those on Earth, sometimes encircling the planet, with wind speeds as high as 60 miles per hour (97 km/h). However, given Mars' much lower atmospheric pressure (roughly 1% that of Earth's), the intensity of Mars storms could never reach the kind of hurricane-force winds that are experienced on Earth.[19] Martian dust storms are formed when solar heating warms the Martian atmosphere and causes the air to move, lifting dust off the ground. The chance for storms is increased when there are great temperature variations like those seen at the equator during the Martian summer.[20]
(Mars Climate Sounder; Mars Reconnaissance Orbiter)
(1:38; animation; 30 October 2018; file description)
A study from 2008 finds that the initial saltation of sand particles induces a static electric field by friction. Saltating sand acquires a negative charge relative to the ground which in turn loosens more sand particles which then begin saltating. This process has been found to double the number of particles predicted by previous theories.[3]
Particles become loosely held mainly due to a prolonged drought or arid conditions, and high wind speeds. Gust fronts may be produced by the outflow of rain-cooled air from an intense thunderstorm. Or, the wind gusts may be produced by a dry cold front: that is, a cold front that is moving into a dry air mass and is producing no precipitation—the type of dust storm which was common during the Dust Bowl years in the U.S. Following the passage of a dry cold front, convective instability resulting from cooler air riding over heated ground can maintain the dust storm initiated at the front.
In desert areas, dust and sand storms are most commonly caused by either thunderstorm outflows, or by strong pressure gradients which cause an increase in wind velocity over a wide area. The vertical extent of the dust or sand that is raised is largely determined by the stability of the atmosphere above the ground as well as by the weight of the particulates. In some cases, dust and sand may be confined to a relatively-shallow layer by a low-lying temperature inversion. In other instances, dust (but not sand) may be lifted as high as 20,000 feet (6,100 m) high.
Drought and wind contribute to the emergence of dust storms, as do poor farming and grazing practices by exposing the dust and sand to the wind.
One poor farming practice which contributes to dust storms is dryland farming. Particularly poor dryland farming techniques are intensive tillage or not having established cropsor cover crops when storms strike at particularly vulnerable times prior to revegetation.[4] In a semi-arid climate, these practices increase susceptibility to dust storms. However, soil conservation practices may be implemented to control wind erosion.
https://en.wikipedia.org/wiki/Dust_storm
In geology, saltation (from Latin saltus, "leap") is a specific type of particle transport by fluids such as wind or water. It occurs when loose materials are removed from a bed and carried by the fluid, before being transported back to the surface. Examples include pebble transport by rivers, sand drift over desert surfaces, soil blowing over fields, and snow drift over smooth surfaces such as those in the Arctic or Canadian Prairies.[citation needed]
https://en.wikipedia.org/wiki/Saltation_(geology)
Asian Dust (also yellow dust, yellow sand, yellow wind or China dust storms) is a meteorological phenomenon that affects much of East Asia year-round and especially during the spring months. The dust originates in China, the deserts of Mongolia, and Kazakhstan where high-speed surface winds and intense dust storms kick up dense clouds of fine, dry soil particles. These clouds are then carried eastward by prevailing winds and pass over China, North and South Korea, and Japan, as well as parts of the Russian Far East. Sometimes, the airborne particulates are carried much further, in significant concentrations which affect air quality as far east as the United States.
Since the turn of the 21st century, coinciding with the rapid industrialization of China, yellow dust has become a serious health problem due to the increase of industrial pollutants contained in the dust. Intensified desertification due to deforestation has been causing longer and more frequent occurrences. The issue has been exacerbated in the last few decades when the Aral Sea of Kazakhstan and Uzbekistan started drying up due to the diversion of the Amu River and Syr River following a Soviet agricultural program to irrigate Central Asian deserts, mainly for cotton plantations.
Some of the earliest written records of dust storm activity are recorded in ancient Chinese literature.[2] It is believed that the earliest Chinese dust storm record was found in the Zhu Shu Ji Nian (Chinese: 竹书纪年; English: the Bamboo Annals).[3] The record said: in the fifth year of Di Xin (1150 BC, Di Xin was the Era Name of the King Di Xin of Shang Dynasty), it rained dust at Bo (Bo is a place in Henan Province in China; in Classical Chinese: 帝辛五年,雨土于亳).
The first known record of an Asian Dust event in Korea was in 174 AD during the Silla Dynasty.[4] The dust was known as "Uto (우토, 雨土)", meaning 'Raining Dirt/Earth', and was believed at the time to be the result of an angry god sending down dust instead of rain or snow. Specific records referring to Asian Dust events in Korea also exist from the Baekje, Goguryeo, and Joseon periods.
Composition[edit]
An analysis of Asian Dust clouds conducted in China in 2001 found that they contain high concentrations of silicon (24–32%), aluminium (5.9–7.4%), calcium (6.2–12%), and iron. Numerous toxic substances were also found, including mercury and cadmium from coal burning.
People further from the source of the dust are more often exposed to nearly invisible, fine dust particles that they can unknowingly inhale deep into their lungs, as coarse dust is too big to be deeply inhaled.[5] After inhalation, these particles can cause long term scarring of lung tissue and induce cancer and lung disease.
Sulfur (an acid rain component), soot, ash, carbon monoxide, and other toxic pollutants including heavy metals (such as mercury, cadmium, chromium, arsenic, lead, zinc, copper) and other carcinogens, often accompany the dust storms, along with viruses, bacteria, fungi, pesticides, antibiotics, asbestos, herbicides, plastic ingredients, combustion products and hormone-mimicking phthalates. Though scientists had known that intercontinental dust plumes can ferry bacteria and viruses, "most people had assumed that the [sun's] ultraviolet light would sterilize these clouds," says microbiologist Dale W. Griffin, "We now find that isn't true."[5]
Research done in 2014 found that yellow dust consists of fine dust and ultrafine dust particles.[6] Fine dust consists of fine particular matter (PM). Particles smaller than 10 µm in diameter are classified as fine PM (PM10), while particles smaller than 2.5 µm in diameter are classified as ultrafine PM (PM2.5). Both fine and ultrafine dust particles impose dangers to health. Fine dust particles are small enough to penetrate deep into the lung alveoli. Ultrafine dust particles are so small that they also penetrate into the blood or lymphatic system through the lungs. Once in the bloodstream, ultrafine particles can even reach the brain or fetal organs.[6]
Asian dust is not a new phenomenon. Historically, there have been records of Asian dust occurrences as early as 1150 B.C. in China and 174 A.D. in Korea.[2][3][4] However, official weather data show a stark increase in its severity and frequency.
In the last half century, the number of days with reports of Asian dust has increased five-fold.[19] According to an analysis on data from Korea Meteorological Administration (KMA), the average number of days with Asian dust in a given year was about two in the 1960s. However, this number has increased to 11 in 2000s. In 1960s and 1970s, each decade had 3 years that were Asian-dust free. However, starting from 2000s, there has not been a single year without Asian dust.[19] In just four months of 2018, Gyeonggi Provinceof South Korea issued 42 dust warnings and advisories, which has increased from 36 in the same period in 2017.[12] This reflects the increase in average dust concentration level from 132.88 ppm (parts per million) in 2017 to 149 ppm in 2018. The situation is worsening since the dust particles are staying in the air longer. The average duration has increased from 16.3 hours to 19.8 hours in the last two years.[20]
| Yellow Dust | |
|---|---|
 Dust clouds leaving mainland China and traveling toward Korea and Japan. |
https://en.wikipedia.org/wiki/Asian_Dust
The Indian Ocean brown cloud or Asian brown cloud is a layer of air pollution that recurrently covers parts of South Asia, namely the northern Indian Ocean, India, and Pakistan.[1][2] Viewed from satellite photos, the cloud appears as a giant brown stain hanging in the air over much of South Asia and the Indian Ocean every year between January and March, possibly also during earlier and later months. The term was coined in reports from the UNEP Indian Ocean Experiment (INDOEX).[3]
The term atmospheric brown cloud is used for a more generic context not specific to the Asian region.[4]
https://en.wikipedia.org/wiki/Asian_brown_cloud
Arctic haze is the phenomenon of a visible reddish-brown springtime haze in the atmosphere at high latitudes in the Arctic due to anthropogenic[1] air pollution. A major distinguishing factor of Arctic haze is the ability of its chemical ingredients to persist in the atmosphere for significantly longer than other pollutants. Due to limited amounts of snow, rain, or turbulent air to displace pollutants from the polar air mass in spring, Arctic haze can linger for more than a month in the northern atmosphere.
https://en.wikipedia.org/wiki/Arctic_haze
The Southeast Asian haze is a fire-related large-scale air pollution problem that occurs regularly. Generally, it is worst between July and October.[1] These haze events have caused adverse health and economic impact on Brunei Darussalam, in Indonesia, Malaysia, Singapore and, to a lesser degree, the Philippines and Thailand.[2][3] The problem flares up every dry season, in varying degrees.[4] Transboundary haze in Southeast Asia has been recorded since 1972.[5]
The haze is largely caused by illegal agricultural fires due to industrial-scale slash-and-burn practices in Indonesia, especially from the provinces of South Sumatra and Riau in Indonesia's Sumatra island, and Kalimantan on Indonesian Borneo. Burned land can be sold at a higher price illegally, and eventually used for activities including oil palm and pulpwood production. Burning is also cheaper and faster compared to cutting and clearing using excavators or other machines.[6][7]
| Southeast Asian haze series |
|---|
 |
Peat (/piːt/), sometimes known as turf (/tɜːrf/), is an accumulation of partially decayed vegetation or organic matter. It is unique to natural areas called peatlands, bogs, mires, moors, or muskegs.[1][2] The peatland ecosystem covers 3.7 million square kilometres (1.4 million square miles)[3] and is the most efficient carbon sink on the planet,[2][4] because peatland plants capture carbon dioxide(CO2) naturally released from the peat, maintaining an equilibrium. In natural peatlands, the "annual rate of biomass production is greater than the rate of decomposition", but it takes "thousands of years for peatlands to develop the deposits of 1.5 to 2.3 m [4.9 to 7.5 ft], which is the average depth of the boreal [northern] peatlands",[2] which store around 415 gigatonnes (Gt) of carbon (about 46 times 2019 global CO2 emissions).[3] Globally, peat stores up to 550 Gt of carbon, 42% of all soil carbon, which exceeds the carbon stored in all other vegetation types, including the world's forests.[5] Across the world, peat covers just 3% of the land’s surface, but stores one-third of the Earth’s soil carbon.[6] Sphagnum moss, also called peat moss, is one of the most common components in peat, although many other plants can contribute. The biological features of sphagnum mosses act to create a habitat aiding peat formation, a phenomenon termed 'habitat manipulation'.[7] Soils consisting primarily of peat are known as histosols. Peat forms in wetlandconditions, where flooding or stagnant water obstructs the flow of oxygen from the atmosphere, slowing the rate of decomposition.[8]Peat properties such as organic matter content and saturated hydraulic conductivity can exhibit high spatial heterogeneity. [9]
Peatlands, particularly bogs, are the primary source of peat;[10] although less-common wetlands including fens, pocosins, and peat swamp forests also deposit peat. Landscapes covered in peat are home to specific kinds of plants including Sphagnum moss, ericaceous shrubs, and sedges (see bog for more information on this aspect of peat). Because organic matter accumulates over thousands of years, peat deposits provide records of past vegetation and climate by preserving plant remains, such as pollen. This allows the reconstruction of past environments and the study of changes in land use.[11]
Peat is used by gardeners and for horticulture in certain parts of the world,[12] but this is being banned in some places.[13] By volume, there are about 4 trillion cubic metres of peat in the world.[14] Over time, the formation of peat is often the first step in the geological formation of fossil fuels such as coal, particularly low-grade coal such as lignite.[15]
Peat is not a renewable source of energy, due to its extraction rate in industrialized countries far exceeding its slow regrowth rate of 1 mm (0.04 in) per year,[16] and as it is also reported that peat regrowth takes place only in 30–40% of peatlands.[17] Centuries of burning and draining of peat by humans has released a significant amount of CO
2 into the atmosphere,[18] and much peatland restoration is needed to help limit climate change.[19]


Peat forms when plant material does not fully decay in acidic and anaerobic conditions. It is composed mainly of wetland vegetation: principally bog plants including mosses, sedges, and shrubs. As it accumulates, the peat holds water. This slowly creates wetter conditions that allow the area of wetland to expand. Peatland features can include ponds, ridges, and raised bogs.[10] The characteristics of some bog plants actively promote bog formation. For example, sphagnum mosses actively secrete tannins, which preserve organic material. Sphagnum also have special water-retaining cells, known as hyaline cells, which can release water ensuring the bogland remains constantly wet which helps promote peat production.[20]
Most modern peat bogs formed 12,000 years ago in high latitudes after the glaciers retreated at the end of the last ice age.[21] Peat usually accumulates slowly at the rate of about a millimetre per year.[16] The estimated carbon content is 415 Gt (4.57×1011 short tons; 4.08×1011 long tons) (northern peatlands),[3] 50 Gt (5.5×1010 short tons; 4.9×1010 long tons) (tropical peatlands) and 15 Gt (1.7×1010 short tons; 1.5×1010 long tons) (South America).[22]
Peat material is either fibric, hemic, or sapric. Fibric peats are the least decomposed and consist of intact fibre. Hemic peats are partially decomposed and sapric are the most decomposed.[23]
Phragmites peat are composed of reed grass, Phragmites australis, and other grasses. It is denser than many other types of peat.
Engineers may describe a soil as peat which has a relatively high percentage of organic material. This soil is problematic because it exhibits poor consolidation properties – it cannot be easily compacted to serve as a stable foundation to support loads, such as roads or buildings.


| Investigation and instrumentation |
| ||||||
|---|---|---|---|---|---|---|---|
| Soil |
| ||||||
| Structures (Interaction) |
| ||||||
| Mechanics |
| ||||||
| Numerical analysis software | |||||||
| Related fields | |||||||
Acid sulfate soils are naturally occurring soils, sediments or organic substrates (e.g. peat) that are formed under waterlogged conditions. These soils contain iron sulfideminerals (predominantly as the mineral pyrite) or their oxidation products. In an undisturbed state below the water table, acid sulfate soils are benign. However, if the soils are drained, excavated or exposed to air by a lowering of the water table, the sulfides react with oxygen to form sulfuric acid.[1]
Release of this sulfuric acid from the soil can in turn release iron, aluminium, and other heavy metals and metalloids (particularly arsenic) within the soil. Once mobilized in this way, the acid and metals can create a variety of adverse impacts: killing vegetation, seeping into and acidifying groundwater[2][3] and surface water bodies,[4][5] killing fish and other aquatic organisms, and degrading concrete and steel structures to the point of failure.[1]
https://en.wikipedia.org/wiki/Acid_sulfate_soil
https://en.wikipedia.org/wiki/Category:Peat-fired_power_stations
In both the World Reference Base for Soil Resources (WRB) and the USDA soil taxonomy, a Histosol is a soil consisting primarily of organic materials. They are defined as having 40 centimetres (16 in) or more of organic soil material in the upper 80 centimetres (31 in). Organic soil material has an organic carbon content (by weight) of 12 to 18 percent, or more, depending on the clay content of the soil. These materials include muck (sapric soil material), mucky peat (hemic soil material), or peat (fibricsoil material). Aquic conditions or artificial drainage are required.[1] Typically, Histosols have very low bulk density and are poorly drained because the organic matter holds water very well. Most are acidic and many are very deficient in major plant nutrientswhich are washed away in the consistently moist soil.
https://en.wikipedia.org/wiki/Histosol
Arctic methane release is the release of methane from seas and soils in permafrost regions of the Arctic. While it is a long-term natural process, methane release is exacerbated by global warming. This results in negative effects, as methane is itself a powerful greenhouse gas.
https://en.wikipedia.org/wiki/Arctic_methane_emissions
Slash-and-burn agriculture is a farming method that involves the cutting and burning of plants in a forest or woodland to create a field called a swidden. The method begins by cutting down the trees and woody plants in an area. The downed vegetation, or "slash", is then left to dry, usually right before the rainiest part of the year. Then, the biomass is burned, resulting in a nutrient-rich layer of ash which makes the soil fertile, as well as temporarily eliminating weed and pest species. After about three to five years, the plot's productivity decreases due to depletion of nutrients along with weed and pest invasion, causing the farmers to abandon the field and move over to a new area. The time it takes for a swidden to recover depends on the location and can be as little as five years to more than twenty years, after which the plot can be slashed and burned again, repeating the cycle.[1][2] In Bangladesh and India, the practice is known as jhum or jhoom.[3][4][5]
The Cycliophora are a platyzoan phylum, based on a single genus Symbion. They are so different from other animals that they were put in their own phylum. They were discovered in 1995, and are the most recent new phylum.[1] So far, three species have been found.
They live on the bodies of cold-water lobsters. They are microscopic: the feeding stage is about 0.3 mm long, and 0.1 mm wide.[2]
Their life style is commensal, (a form of symbiosis) – they feed on the leftovers from the lobster's own meals.[3]
https://simple.wikipedia.org/wiki/Cycliophora
above. D12 - My Band ft. Cameo (Official Music Video)
1930, Puttin' On The Ritz, Phil Spitalny Orch. Hi Def 78RPM .wmv



/GettyImages-604395517-ab3c2b6598b04fc2a9faee6def034b39.jpg)




































Trichoplax adhaerens is one of the three named species in the phylum Placozoa. The others are Hoilungia hongkongensis and Polyplacotoma mediterranea. The Placozoa is a basal group of multicellular animals (metazoa). Trichoplax are very flat organisms around a millimetre in diameter, lacking any organs or internal structures. They have two cellular layers: the top epitheloid layer is made of ciliated "cover cells" flattened toward the outside of the organism, and the bottom layer is made up of cylinder cells that possess cilia used in locomotion, and gland cells that lack cilia.[2] Between these layers is the fibre syncytium, a liquid-filled cavity strutted open by star-like fibres.
Trichoplax feed by absorbing food particles—mainly microbes—with their underside. They generally reproduce asexually, by dividing or budding, but can also reproduce sexually. Though Trichoplax has a small genome in comparison to other animals, nearly 87% of its 11,514 predicted protein-coding genes are identifiably similar to known genes in other animals.
| Trichoplax | |
|---|---|
 | |
| Light microscope image of Trichoplax(specimen ca. 0.5 mm across) | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Placozoa |
| Family: | Trichoplacidae Bütschli & Hatschek, 1905 |
| Genus: | Trichoplax Schulze, 1883 [1] |
| Species: | T. adhaerens |
| Binomial name | |
| Trichoplax adhaerens Schulze, 1883 | |
| Synonyms | |
| |
Trichoplax was discovered in 1883 by the German zoologist Franz Eilhard Schulze, in a seawater aquarium at the Zoological Institute in Graz, Austria. The generic name is derived from the classical Greek θρίξ (thrix), "hair", and πλάξ (plax), "plate". The specific epithet adhaerens comes from Latin "adherent", reflecting its propensity to stick to the glass slides and pipettes used in its examination.[3]
Although from the very beginning most researchers who studied Trichoplax in any detail realized that it had no close relationship to other animal phyla, the zoologist Thilo Krumbach published a hypothesis that Trichoplax is a form of the planula larva of the anemone-like hydrozoan Eleutheria krohni in 1907. Although this was refuted in print by Schulze and others, Krumbach's analysis became the standard textbook explanation, and nothing was printed in zoological journals about Trichoplax until the 1960s. In the 1960s and 1970s a new interest among researchers led to acceptance of Placozoa as a new animal phylum. Among the new discoveries was study of the early phases of the animals' embryonic development and evidence that the animals that people had been studying are adults, not larvae. This newfound interest also included study of the organism in nature (as opposed to aquariums).[4]
https://en.wikipedia.org/wiki/Trichoplax
A desmosome (/ˈdɛzməˌsoʊm/;[1][2] "binding body"), also known as a macula adherens (plural: maculae adherentes) (Latin for adhering spot), is a cell structure specialized for cell-to-cell adhesion. A type of junctional complex, they are localized spot-like adhesions randomly arranged on the lateral sides of plasma membranes. Desmosomes are one of the stronger cell-to-cell adhesion types and are found in tissue that experience intense mechanical stress, such as cardiac muscle tissue, bladder tissue, gastrointestinal mucosa, and epithelia.[3]
| Desmosome | |
|---|---|
 A desmosome. |

Arrhythmogenic cardiomyopathy[edit]
Mutations within the desmosome are the main cause of arrhythmogenic cardiomyopathy (ACM), a life-threatening disease caused by mutations usually in desmoglein 2, but sometimes in desmocollin 2. It often afflicts individuals between 20-50 years, and has been publicly known as a cause of death in young athletes, although the majority of sudden deaths do not occur in close connection to physical activity. The current incidence within the population is accepted as 1/10,000 however it is thought that 1/200 may have a mutation that may predispose to ACM.[8] Symptoms of ACM include fainting, shortness of breath, and heart palpitations and the condition is treated by implanting a small defibrillator device.
Blisters[edit]
Blistering diseases such as pemphigus vulgaris (PV) and pemphigus foliaceus (PF) are autoimmune diseases in which auto-antibodies target desmogleins. PV is caused by circulating autoantibodies (IgG) that target Dsg3 (Desmoglein 3) and sometimes Dsg1. PV is manifested by suprabasal acantholysis, or blisters in the mucous membrane and blisters in the epidermis. PF patients have autoantibodies that target Dsg1 with superficial blisters on the epidermis with no mucous membrane issues. Both disease result in a loss of keratinocyte adhesion. Pemphigus can also be caused by a bacterial infection: bullous impetigo is an infection caused by a staphylococcus bacterium that releases a toxin that cleaves the Dsg1 extracellular domain.
Similar symptoms occur with Hailey–Hailey disease, though the cause is not autoimmune but genetic. A haploinsufficiency of the ATP2C1 gene located on chromosome 3, which encodes the protein hSPCA1, causes malformation of the desmosomes. Desmoglein 1 haploinsufficiency leads to striate palmoplantar keratoderma, a disease which causes extreme thickening of the epidermis. Loss of desmoglein 4 leads to defective hair-follicle differentiation.[3]
Epidermolysis bullosa simplex is an epidermal blistering disease caused by mutations in genes coding for keratin 5 and 14, which attach to desmoplakin. This disease manifests as rupture of the basal epidermis when stress is applied. Ectodermal dysplasia or skin fragility syndrome is caused by plakophillin 1 mutations. This is manifested by detachment of intermediate filaments and desmoplakin from the desmosome.[4]
History[edit]
The desmosome was first discovered by Giulio Bizzozero, an Italian pathologist.[3] He named these "dense nodules" the "nodes of Bizzozero". In 1920, the term "desmosome" was originated by Josef Schaffer. The first combining form, desmo-, New Latin from Greek desmos, bond, carries meaning of binding or bonding things together. Combined with -some, which comes from soma, body, it thus makes a desmosome a "binding body".
https://en.wikipedia.org/wiki/Desmosome
Arrhythmogenic cardiomyopathy (ACM), arrhythmogenic right ventricular dysplasia (ARVD), or arrhythmogenic right ventricular cardiomyopathy (ARVC), is an inherited heart disease.[1]
ACM is caused by genetic defects of the parts of heart muscle (also called myocardium or cardiac muscle) known as desmosomes, areas on the surface of heart muscle cells which link the cells together. The desmosomes are composed of several proteins, and many of those proteins can have harmful mutations.
The disease is a type of non-ischemic cardiomyopathy that primarily involves the right ventricle, though cases of exclusive left ventricular disease have been reported. It is characterized by hypokinetic areas involving the free wall of the ventricle, with fibrofatty replacement of the myocardium, with associated arrhythmias often originating in the right ventricle. The nomenclature ARVD is currently thought to be inappropriate and misleading as ACM does not involve dysplasia of the ventricular wall. Cases of ACM originating from the left ventricle led to the abandonment of the name ARVC.
ACM can be found in association with diffuse palmoplantar keratoderma, and woolly hair, in an autosomal recessive condition called Naxos disease, because this genetic abnormality can also affect the integrity of the superficial layers of the skin most exposed to pressure stress.[2]:513[3]
ACM is an important cause of ventricular arrhythmias in children and young adults. It is seen predominantly in males, and 30–50% of cases have a familial distribution.
| Other names | arrhythmogenic right ventricular cardiomyopathy (ARVC), arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C), right ventricular dysplasia |
|---|---|
 | |
| Typical micro-histologic features of ARVC/D. Ongoing myocyte death (upper) with early fibrosis and adipocyte infiltration (lower). | |
| Specialty | Cardiology |
Naxos disease (also known as "diffuse non-epidermolytic palmoplantar keratoderma with woolly hair and cardiomyopathy,"[1] "diffuse palmoplantar keratoderma with woolly hair and arrhythmogenic right ventricular cardiomyopathy, first described on the island of Naxos by Nikos Protonotarios,"[1] and "Naxos disease"[1]) is a cutaneous condition characterized by a palmoplantar keratoderma.[1] The prevalence of the syndrome is up to 1 in every 1000 people in the Greek islands.[2]
It has been associated with mutations in the genes encoding the proteins desmoplakin, plakoglobin, desmocollin-2, and SRC-interacting protein (SIP).[3][4] A variation of Naxos syndrome is known as Carvajal syndrome.[2]
| Naxos disease | |
|---|---|
| Other names | Diffuse non-epidermolytic palmoplantar keratoderma with woolly hair and cardiomyopathy |
 | |
| Cutaneous phenotype of Naxos disease: woolly hair (A), palmar (B) and plantar (C) keratoses. | |
Epidermolysis bullosa simplex (EBS), is a disorder resulting from mutations in the genes encoding keratin 5 or keratin 14.[1]:598[2]
Blister formation of EBS occurs at the dermoepidermal junction. Sometimes EBS is called epidermolytic.[citation needed]
https://en.wikipedia.org/wiki/Epidermolysis_bullosa_simplex
Epidermolysis bullosa (EB) is a group of rare medical conditions that result in easy blistering of the skin and mucous membranes.
Epidermolysis bullosa simplex[edit]
Epidermolysis bullosa simplex (EBS) is a form of EB that causes blisters at the site of rubbing. It typically affects the hands and feet, and is typically inherited in an autosomal dominant manner, affecting the keratin genes KRT5 and KRT14. Therefore, there is a failure in keratinisation, which affects the integrity and the ability of the skin to resist mechanical stresses.[citation needed]
Junctional epidermolysis bullosa[edit]
Junctional epidermolysis bullosa (JEB) is an inherited disease affecting laminin and collagen. This disease is characterised by blister formation within the lamina lucida of the basement membrane zone[12]:599 and is inherited in an autosomal recessive manner. It also presents with blisters at the site of friction, especially on the hands and feet, and has variants that can occur in children and adults. Less than one person per million people is estimated to have this form of EB.[13]
Dystrophic epidermolysis bullosa[edit]
Dystrophic epidermolysis bullosa (DEB) is an inherited variant affecting the skin and other organs. DEB is caused by genetic defects (or mutations) within the human COL7A1 gene encoding the protein type VII collagen (collagen VII).[14] DEB-causing mutations can be either autosomal dominant or autosomal recessive. Epidermis bullosa pruriginosaand albopapuloid epidermolysis bullosa (Pasini's disease) are rare subtypes of this disease.[15]
Other genetic[edit]
| OMIM | Name | Locus | Gene |
|---|---|---|---|
| 609638 | epidermolysis bullosa, lethal acantholytic | 6p24 | DSP |
Epidermolysis bullosa acquisita[edit]
Acral peeling[edit]
Recent research has focused on changing the mixture of keratins produced in the skin. There are 54 known keratin genes—of which 28 belong to the type I intermediate filament genes and 26 to type II—which work as heterodimers. Many of these genes share substantial structural and functional similarity, but they are specialized to cell type and/or conditions under which they are normally produced. If the balance of production could be shifted away from the mutated, dysfunctional keratin gene toward an intact keratin gene, symptoms could be reduced. For example, sulforaphane, a compound found in broccoli, was found to reduce blistering in a mouse model to the point where affected pups could not be identified visually, when injected into pregnant mice (5 μmol/day = 0.9 mg) and applied topically to newborns (1 μmol/day = 0.2 mg in jojoba oil).[16]
As of 2008 clinical research at the University of Minnesota has explored allogeneic bone marrow transplantation for RD and junctional EB, treating a 2-year-old child who is one of 2 brothers with EB. A second transplant has also been performed on the child's older brother, and a third transplant is scheduled for a California baby. A clinical trial is planned for 30 subjects.[17] However, the immune suppression that bone marrow transplantation requires causes a risk of serious infections with large scale blisters and skin erosion.[18]Indeed, at least four people have died in the course of either preparation for or institution of bone marrow transplantation for EB, out of only a small group of patients treated so far.[18] The mechanism of action of this therapy is unclear as hematopoietic stem cells are not thought to contribute to epithelial lineages. Rather, it is speculated that cross-correction from tissue-resident graft-derived immune cells contributes to the observed clinical benefit.[19]
A pilot study performed in 2015 suggests that systemic granulocyte-colony stimulating factor (G-CSF) may promote increased wound healing in people with dystrophic EB.[20]Transplanting skin derived from genetically modified stem cells onto the wound surfaces has been studied with a report of improvements in one person.[21]
https://en.wikipedia.org/wiki/Epidermolysis_bullosa
Ref. syncytin syntactin matrix matricing adhesion complex complexing level dimension primary secondary tertiary quaternary protein disease prion virus genetic organism junction cell junction
human foamy virus, matricial collapse/dysfunction/det/etc. cell cycle interspace fluid etc.
Neonatal ichthyosis–sclerosing cholangitis syndrome (also known as "NISCH syndrome"[1] and "ichthyosis–sclerosing cholangitis syndrome"[1]) is a cutaneous condition caused by mutations in the Claudin 1 gene.[1]
https://en.wikipedia.org/wiki/Neonatal_ichthyosis–sclerosing_cholangitis_syndrome
Ichthyosis vulgaris (also known as "autosomal dominant ichthyosis"[1] and "Ichthyosis simplex"[1]) is a skin disorder causing dry, scaly skin. It is the most common form of ichthyosis,[2]:486 affecting around 1 in 250 people.[3] For this reason it is known as common ichthyosis. It is usually an autosomal dominant inherited disease (often associated with filaggrin), although a rare non-heritable version called acquired ichthyosis exists.[4]:560
| Ichthyosis vulgaris | |
|---|---|
 | |
| Ichthyosis vulgaris #1 (top-left) | |
| Specialty | Medical genetics |
https://en.wikipedia.org/wiki/Ichthyosis_vulgaris
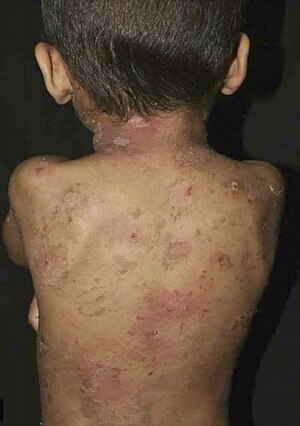
Epidermolytic ichthyosis (EI), also known as bullous epidermis ichthyosis (BEI), epidermolytic hyperkeratosis(EHK), bullous congenital ichthyosiform erythroderma (BCIE),[1] bullous ichthyosiform erythroderma[2]:482 or bullous congenital ichthyosiform erythroderma Brocq,[3] is a rare and severe form of ichthyosis this skin disease affects around 1 in 300,000 people.
The condition is caused by a genetic mutation, so it cannot be completely cured without some form of gene therapy.
While some research has been done into possible gene therapy treatments, the work hasn't yet been successfully developed to the stage where it can be routinely given to patients.
The condition involves the clumping of keratin filaments.[4][5]:562
| Epidermolytic Ichthyosis (EI) | |
|---|---|
| Other names | Bullous epidermis ichthyosis |
 | |
| Specialty | Medical genetics |
The challenge has always been how to deliver the siRNA using a topical method or retroviral vectors and ex vivo gene transfer.[10] In 2011/12 a team at Northwestern University claim to have solved the topical delivery of siRNA dilemma. Personalized siRNA can be delivered in a commercial moisturizer or phosphate-buffered saline, and do not require barrier disruption or transfection agents, such as liposomes, peptides, or viruses. "Topical application of nucleic acids offers many potential therapeutic advantages for suppressing genes in the skin, and potentially for systemic gene delivery. However, the epidermal barrier typically precludes entry of gene-suppressing therapy unless the barrier is disrupted. We now show that spherical nucleic acid nanoparticle conjugates (SNA-NCs), gold cores surrounded by a dense shell of highly oriented, covalently immobilized siRNA, freely penetrate almost 100% of keratinocytes in vitro, mouse skin, and human epidermis within hours after application."[11] This new discovery may soon offer hope to all suffering from mono-genetic diseases such as EHK. This may lead to promising personalized, topically delivered gene therapy of cutaneous tumors, skin inflammation, and dominant negative genetic skin disorders.
[12] UPDATE: OCTOBER 2014 As of late, Paller reports "we are using a new nanotechnology-based technique called 'spherical nucleic acids' (SNAs) to suppress the production of the abnormal keratin 10 gene that is the most common change leading to epidermolytic ichthyosis. We continue to screen candidate SNAs to find a few that clearly suppress the abnormal keratin 10 gene much more than the normal keratin 10 gene. In the meantime, we have developed several tools towards this effort, which can also be used by other researchers. Most recently we've developed a special 'lentivirus reporter construct' in which we can see through changes in fluorescence whether or not our SNA works."
Dr. Paller and her team recently received more good news with regard to progressing their research. "We just received a grant from the National Institutes of Health (NIH) to continue this effort based on our preliminary data collected with FIRST's funding support. FIRST has been instrumental in furthering our research efforts related to ichthyosis," she said.
https://en.wikipedia.org/wiki/Epidermolytic_hyperkeratosis
Ichthyosis is a family of genetic skin disorders characterized by dry, thickened, scaly skin.[1] The more than 20 types of ichthyosis range in severity of symptoms, outward appearance, underlying genetic cause and mode of inheritance (e.g., dominant, recessive, autosomal or X-linked).[2] Ichthyosis comes from the Greek ἰχθύς ichthys, literally "fish", since dry, scaly skin is the defining feature of all forms of ichthyosis.[3]
The severity of symptoms can vary enormously, from the mildest, most common, types such as ichthyosis vulgaris, which may be mistaken for normal dry skin, up to life-threatening conditions such as harlequin-type ichthyosis. Ichthyosis vulgaris accounts for more than 95% of cases.[4]
| hthyosis | |
|---|---|
| Other names | Ichthyoses |
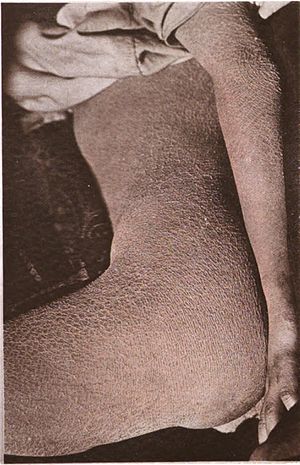 | |
| Ichthyosis is characterized by rough, scaly skin. | |
https://en.wikipedia.org/wiki/Ichthyosis
Lamellar ichthyosis, also known as ichthyosis lamellaris and nonbullous congenital ichthyosis, is a rare inherited skin disorder, affecting around 1 in 600,000 people.
In medicine, the term collodion baby applies to newborns who appear to have an extra layer of skin (known as a collodion membrane) that has a collodion-like quality. It is a descriptive term, not a specific diagnosis or disorder (as such, it is a syndrome).[1]
https://en.wikipedia.org/wiki/Lamellar_ichthyosis
Collodion is a flammable, syrupy solution of nitrocellulose in ether and alcohol. There are two basic types: flexible and non-flexible. The flexible type is often used as a surgical dressing or to hold dressings in place. When painted on the skin, collodion dries to form a flexible nitrocellulose film. While it is initially colorless, it discolors over time. Non-flexible collodion is often used in theatrical make-up.
In 1851, Frederick Scott Archer, an Englishman, discovered that collodion could be used as an alternative to egg white (albumen) on glass photographic plates.[5] Collodion reduced the exposure time necessary for making an image. This method became known as the 'wet-plate collodion' or 'wet collodion' method. Collodion was relatively grainless and colorless, and allowed for one of the first high-quality duplication processes, also known as negatives. This process also produced two types of positives: the ambrotype and the tintype (also known as ferrotype).
The process required great skill and included the following steps:
- Clean the glass plate (extremely well)
- In the light, pour "salted" (iodide, bromide) collodion onto the glass plate, tilting it so it reaches each corner. The excess is poured back into the bottle.
- Take the plate into a darkroom or orange tent (the plate is sensitive only to blue light) and immerse the plate in a silver nitratesensitising bath (for 3–5 minutes)
- Lift the plate out of the bath, drain and wipe the back, load it into a plate holder and protect from light with a dark slide.
- Load the plate holder into the camera, withdraw the dark slide and expose the plate (can range from less than a second to several minutes)
- Develop the plate (using a ferrous sulfate based developer)
- Fix the plate (with potassium cyanide or sodium thiosulfate)
All of this was done in a matter of minutes, and some of the steps in (red) safelight conditions, which meant that the photographer had to carry the chemicals and a portable darkroom with him wherever he went. After these steps the plate needed rinsing in fresh water. Finally, the plate was dried and varnished using a varnish made from sandarac, alcohol and lavender oil.
Dark tents to be used outdoors consisted of a small tent that was tied around the photographer's waist. Otherwise a wheelbarrow or a horse and covered wagon were used.
https://en.wikipedia.org/wiki/Collodion
Epidermolytic ichthyosis (EI), also known as bullous epidermis ichthyosis (BEI), epidermolytic hyperkeratosis(EHK), bullous congenital ichthyosiform erythroderma (BCIE),[1] bullous ichthyosiform erythroderma[2]:482 or bullous congenital ichthyosiform erythroderma Brocq,[3] is a rare and severe form of ichthyosis this skin disease affects around 1 in 300,000 people.
The condition is caused by a genetic mutation, so it cannot be completely cured without some form of gene therapy.
While some research has been done into possible gene therapy treatments, the work hasn't yet been successfully developed to the stage where it can be routinely given to patients.
The condition involves the clumping of keratin filaments.[4][5]:562
https://en.wikipedia.org/wiki/Epidermolytic_hyperkeratosis
Measles is a highly contagious infectious disease caused by measles virus.[11][3][12] Symptoms usually develop 10–12 days after exposure to an infected person and last 7–10 days.[7][8] Initial symptoms typically include fever, often greater than 40 °C (104 °F), cough, runny nose, and inflamed eyes.[3][4] Small white spots known as Koplik's spots may form inside the mouth two or three days after the start of symptoms.[4] A red, flat rash which usually starts on the face and then spreads to the rest of the body typically begins three to five days after the start of symptoms.[4] Common complications include diarrhea (in 8% of cases), middle ear infection (7%), and pneumonia (6%).[5] These occur in part due to measles-induced immunosuppression.[6] Less commonly seizures, blindness, or inflammation of the brain may occur.[5][7] Other names include morbilli, rubeola, red measles, and English measles.[1][2] Both rubella, also known as German measles, and roseola are different diseases caused by unrelated viruses.[13]
| Measles | |
|---|---|
| Other names | Morbilli, rubeola, red measles, English measles[1][2] |
 | |
| A child showing a day-four measles rash | |
| Complications | Pneumonia, seizures, encephalitis, subacute sclerosing panencephalitis, immunosuppression, hearing loss, blindness[5][6] |
|---|
Once the measles virus gets onto the mucosa, it infects the epithelial cells in the trachea or bronchi.[56][57] Measles virus uses a protein on its surface called hemagglutinin (H protein), to bind to a target receptor on the host cell, which could be CD46, which is expressed on all nucleated human cells, CD150, aka signaling lymphocyte activation molecule or SLAM, which is found on immune cells like B or T cells, and antigen-presenting cells, or nectin-4, a cellular adhesion molecule.[56][58] Once bound, the fusion, or F protein helps the virus fuse with the membrane and ultimately get inside the cell.[56]
As the virus is a single-stranded negative-sense RNA virus, it includes the enzyme RNA-dependent RNA polymerase(RdRp) which is used to transcribe its genome into a positive-sense mRNA strand.[56]
After entering a cell, it is ready to be translated into viral proteins, wrapped in the cell's lipid envelope, and sent out of the cell as a newly made virus.[59] Within days, the measles virus spreads through local tissue and is picked up by dendritic cells and alveolar macrophages, and carried from that local tissue in the lungs to the local lymph nodes.[56][57]From there it continues to spread, eventually getting into the blood and spreading to more lung tissue, as well as other organs like the intestines and the brain.[26][56] Functional impairment of the infected dendritic cells by the measles virus is thought to contribute to measles-induced immunosuppression.[6]
https://en.wikipedia.org/wiki/Measles
https://en.wikipedia.org/wiki/Measles
Signaling lymphocytic activation molecule 1 is a protein that in humans is encoded by the SLAMF1 gene.[5][6]Recently SLAMF1 has also been designated CD150 (cluster of differentiation 150).
SLAMF1 belongs to the signaling lymphocytic activation molecule family. The interaction of SLAMF1 promoter and enhancers with the Early B-cell factor 1 (EBF1) is required for the expression of SLAMF1 gene in B cells. STAT6, IRF4, and NF-kB factors involved in the transfer of the signals from the B-cell receptor, its co-receptors and IL-4R, also play important role in the regulation of SLAMF1 expression.[7]
https://en.wikipedia.org/wiki/SLAMF1
Category:Clusters of differentiation
| Wikimedia Commons has media related to Clusters of differentiation. |
In immunology and cell biology, clusters of differentiation are molecules on the cell surface, as recognized by specific sets of antibodies, used to identify the cell type, stage of differentiation and activity of a cell.
Subcategories
This category has the following 2 subcategories, out of 2 total.
Pages in category "Clusters of differentiation"
The following 200 pages are in this category, out of approximately 306 total. This list may not reflect recent changes (learn more).
(previous page) (next page)B
C
- C-C chemokine receptor type 6
- C-C chemokine receptor type 7
- CCR1
- CCR2
- CCR3 (gene)
- CCR4
- CCR5
- CCR8 (gene)
- CCR9
- CD 205
- CD1
- CD1A
- CD1b
- CD1E
- CD2
- CD3 (immunology)
- CD3D
- CD3G
- CD4
- CD4+/CD8+ ratio
- CD5
- CD5 (protein)
- CD6
- CD7
- CD8
- CD8A
- CD11
- CD14
- CD15
- CD16
- CD19
- CD20
- CD23
- CD28
- CD31
- CD32
- CD34
- CD36
- CD37
- CD38
- CD40 (protein)
- CD43
- CD44
- CD46
- CD47
- CD48
- CD53
- Neural cell adhesion molecule
- CD59
- CD63
- CD64 (biology)
- CD68
- CD69
- CD70
- CD72
- CD74
- CD78
- CD79
- CD79A
- CD79B
- CD80
- CD81
- CD82 (gene)
- CD83
- CD84
- CD86
- CD90
- CD93
- CD96
- CD98
- CD99
- CD109
- CD134
- Tissue factor
- CD151
- CD154
- CD155
- CD160
- CD163
- CD177
- CD180
- CD200
- CD226
- CD244
- CD248
- CD276
- CD278
- CD300A
- CD320
- CDCP1
- CDH1 (gene)
- CDH2
- CEACAM1
- CEACAM3
- CEACAM5
- CEACAM6
- CEACAM8
- Choline transporter-like protein 1
- CLEC4M
- CNPY2
- Colony stimulating factor 1 receptor
- Complement receptor 1
- CTLA-4
- CXCR3
- CXCR5
- CXCR6
D
F
G
I
- ICAM2
- ICAM3
- ICOSLG
- IFITM1
- IGLL1
- IGSF2
- IGSF8
- IL2RA
- IL3RA
- IL13RA2
- IL17RA
- IL18R1
- IL18RAP
- Insulin receptor
- Insulin-like growth factor 1 receptor
- Insulin-like growth factor 2 receptor
- Integrin alpha 2b
- Integrin alpha 3
- Integrin alpha 4
- Integrin alpha 5
- Integrin alpha 6
- Integrin alpha L
- Integrin alpha V
- Integrin alpha X
- Integrin beta 1
- Integrin beta 2
- Integrin beta 4
- Interferon gamma receptor 1
- Interleukin 1 receptor, type I
Once the measles virus gets onto the mucosa, it infects the epithelial cells in the trachea or bronchi.[56][57] Measles virus uses a protein on its surface called hemagglutinin (H protein), to bind to a target receptor on the host cell, which could be CD46, which is expressed on all nucleated human cells, CD150, aka signaling lymphocyte activation molecule or SLAM, which is found on immune cells like B or T cells, and antigen-presenting cells, or nectin-4, a cellular adhesion molecule.[56][58] Once bound, the fusion, or F protein helps the virus fuse with the membrane and ultimately get inside the cell.[56]
As the virus is a single-stranded negative-sense RNA virus, it includes the enzyme RNA-dependent RNA polymerase(RdRp) which is used to transcribe its genome into a positive-sense mRNA strand.[56]
After entering a cell, it is ready to be translated into viral proteins, wrapped in the cell's lipid envelope, and sent out of the cell as a newly made virus.[59] Within days, the measles virus spreads through local tissue and is picked up by dendritic cells and alveolar macrophages, and carried from that local tissue in the lungs to the local lymph nodes.[56][57]From there it continues to spread, eventually getting into the blood and spreading to more lung tissue, as well as other organs like the intestines and the brain.[26][56] Functional impairment of the infected dendritic cells by the measles virus is thought to contribute to measles-induced immunosuppression.[6]
Other names include morbilli, rubeola, red measles, and English measles.[1][2]
https://en.wikipedia.org/wiki/Measles
Scarlet fever is a disease resulting from a group A streptococcus (group A strep) infection, also known as Streptococcus pyogenes.[1] The signs and symptoms include a sore throat, fever, headaches, swollen lymph nodes, and a characteristic rash.[1] The rash is red and feels like sandpaper and the tongue may be red and bumpy.[1] It most commonly affects children between five and 15 years of age.[1]
Scarlet fever affects a small number of people who have strep throat or streptococcal skin infections.[1] The bacteria are usually spread by people coughing or sneezing.[1] It can also be spread when a person touches an object that has the bacteria on it and then touches their mouth or nose.[1] The characteristic rash is due to the erythrogenic toxin, a substance produced by some types of the bacterium.[1][4] The diagnosis is typically confirmed by culturing the throat.[1]
As of 2020 there is no vaccine.[5] Prevention is by frequent handwashing, not sharing personal items, and staying away from other people when sick.[1] The disease is treatable with antibiotics, which prevent most complications.[1] Outcomes with scarlet fever are typically good if treated.[3] Long-term complications as a result of scarlet fever include kidney disease, rheumatic heart disease, and arthritis.[1] In the early 20th century, before antibiotics were available, it was a leading cause of death in children.[6][7] An antitoxin was produced before antibiotics; however, it was never made in sufficient quantities, and could not be used to treat any other disease as antibiotics can.
There have been signs of antibiotic resistance, and there have been recent outbreaks in Hong Kong in 2011 and in the UK in 2014, with occurrence rising 68% in the UK in the four years up to 2018. Research published in October 2020 has shown that infection of the bacterium by three viruses has led to stronger strains of the bacterium.[5]
https://en.wikipedia.org/wiki/Scarlet_fever
Dengue fever is a mosquito-borne tropical disease caused by the dengue virus.[1] Symptoms typically begin three to fourteen days after infection.[2] These may include a high fever, headache, vomiting, muscle and joint pains, and a characteristic skin rash.[1][2] Recovery generally takes two to seven days.[1] In a small proportion of cases, the disease develops into a more severe dengue hemorrhagic fever, resulting in bleeding, low levels of blood platelets and blood plasma leakage, or into dengue shock syndrome, where dangerously low blood pressure occurs.[1][2]
Dengue is spread by several species of female mosquitoes of the Aedes genus, principally Aedes aegypti.[1][2] The virus has five serotypes;[7][8] infection with one type usually gives lifelong immunity to that type, but only short-term immunity to the others.[1]Subsequent infection with a different type increases the risk of severe complications.[1] A number of tests are available to confirm the diagnosis including detecting antibodies to the virus or its RNA.[2]
A vaccine for dengue fever has been approved and is commercially available in a number of countries.[4][9] As of 2018, the vaccine is only recommended in individuals who have been previously infected, or in populations with a high rate of prior infection by age nine.[10][5] Other methods of prevention include reducing mosquito habitat and limiting exposure to bites.[1] This may be done by getting rid of or covering standing water and wearing clothing that covers much of the body.[1] Treatment of acute dengue is supportive and includes giving fluid either by mouth or intravenously for mild or moderate disease.[2] For more severe cases, blood transfusion may be required.[2] Paracetamol (acetaminophen) is recommended instead of nonsteroidal anti-inflammatory drugs (NSAIDs) for fever reduction and pain relief in dengue due to an increased risk of bleeding from NSAID use.[2][11][12]
The earliest descriptions of an outbreak date from 1779.[13] Its viral cause and spread were understood by the early 20th century.[14] Dengue has become a global problem since the Second World War and is common in more than 120 countries, mainly in Southeast Asia, South Asia and South America.[5][15][13] About 390 million people are infected a year, about half a million require hospital admission,[1] and approximately 40,000 die.[5][6] In 2019 a significant increase in the number of cases was seen.[16] Apart from eliminating the mosquitos, work is ongoing for medication targeted directly at the virus.[17] It is classified as a neglected tropical disease.[18]
https://en.wikipedia.org/wiki/Dengue_fever
Parvoviruses are a family of animal viruses that constitute the family Parvoviridae. They have linear, single-stranded DNA (ssDNA) genomes that typically contain two genes encoding for a replication initiator protein, called NS1, and the protein the viral capsid is made of. The coding portion of the genome is flanked by telomeres at each end that form into hairpin loops that are important during replication. Parvovirus virions are small compared to most viruses, at 23–28 nanometers in diameter, and contain the genome enclosed in an icosahedral capsid that has a rugged surface.
Parvoviruses enter a host cell by endocytosis, travelling to the nucleus where they wait until the cell enters its replication stage. At that point, the genome is uncoated and the coding portion is replicated. Viral messenger RNA (mRNA) is then transcribed and translated, resulting in NS1 initiating replication. During replication, the hairpins repeatedly unfold, are replicated, and refold to change the direction of replication to progress back and forth along the genome in a process called rolling hairpin replication that produces a molecule containing numerous copies of the genome. Progeny ssDNA genomes are excised from this concatemer and packaged into capsids. Mature virions leave the cell by exocytosis or lysis.
Parvoviruses are believed to be descended from ssDNA viruses that have circular genomes that form a loop because these viruses encode a replication initiator protein that is related to NS1 and have a similar replication mechanism. Another group of viruses called bidnaviruses appear to be descended from parvoviruses. Within the family, three subfamilies, 26 genera, and 126 species are recognized. Parvoviridae is the sole family in the order Piccovirales, which is the sole order in the class Quintoviricetes. This class is assigned to the phylum Cossaviricota, which also includes papillomaviruses, polyomaviruses, and bidnaviruses.
A variety of diseases in animals are caused by parvoviruses. Notably, the canine parvovirus and feline parvovirus cause severe disease in dogs and cats, respectively. In pigs, the porcine parvovirus is a major cause of infertility. Human parvoviruses are less severe, the two most notable being parvovirus B19, which causes a variety of illnesses including Fifth disease in children, and human bocavirus 1, which is a common cause of acute respiratory tract illness, especially in young children. In medicine, recombinant adeno-associated viruses (AAV) have become an important vector for delivering genes to the cell nucleus during gene therapy.
Animal parvoviruses were first discovered in the 1960s, including minute virus of mice, which is frequently used to study parvovirus replication. Many AAVs were also discovered during this time period and research on them over time has revealed their benefit as a form of medicine. The first pathogenic human parvovirus to be discovered was parvovirus B19 in 1974, which became associated with various diseases throughout the 1980s. Parvoviruses were first classified as the genus Parvovirus in 1971 but were elevated to family status in 1975. They take their name from the Latin word parvum, meaning small or tiny, referring to the small size of the virus's virions.
| Parvoviridae | |
|---|---|
 | |
| Electron micrograph of canine parvovirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Quintoviricetes |
| Order: | Piccovirales |
| Family: | Parvoviridae |
| Genera | |
Parvoviruses have linear, single-stranded DNA (ssDNA) genomes that are about 4–6 kilobases (kb) in length. The parvovirus genome typically contains two genes, termed the NS/rep gene and the VP/cap gene.[1] The NS gene encodes the non-structural (NS) protein NS1, which is the replication initiator protein, and the VP gene encodes the viral protein (VP) that the viral capsid is made of. NS1 contains an HUH superfamily endonuclease domain near its N-terminus, containing both site-specific binding activity and site-specific nicking activity, and a superfamily 3 (SF3) helicase domain toward the C-terminus. Most parvoviruses contain a transcriptional activation domain near the C-terminus that upregulates transcription from viral promoters as well as alternate or overlapping open reading frames that encode a small number of supporting proteins involved in different aspects of the viral life cycle.[2]
The coding portion of the genome is flanked at each end by terminal sequences about 116–550 nucleotides (nt) in length that consist of imperfect palindromes folded into hairpin loop structures. These hairpin loops contain most of the cis-acting information required for DNA replication and packaging and act as hinges during replication to change the direction of replication. When the genome is converted to double-stranded forms, replication origin sites are created involving sequences in and adjacent to the hairpins.[2][3]
Genomic DNA strands in mature virions may be positive-sense or negative-sense. This varies from species to species as some have a preference for packaging strands of one polarity, others package varying proportions, and others package both sense strands at equal proportions. These preferences reflect the efficiency with which progeny strands are synthesized, which in turn reflects the efficiency of specific replication origin sites.[2] The 3′-end (usually pronounced "three prime end") of a negative sense strand, and the 5′-end (usually pronounced "five prime end") of a positive sense strand, is called the left end, and the 5′-end of the negative sense strand, and the 3′-end of a positive sense strand, is called the right end.[2][4][5]
Structure[edit]
Parvovirus virions are 23–28 nanometers (nm) in diameter and consist of the genome enclosed inside a capsid that is icosahedralin shape with a rugged surface. The capsid is composed of 60 structurally equivalent polypeptide chains derived from the C-terminal end of a VP protein's sequence, interlocking extensively to form an icosahedron with 60 asymmetric, superficial triangular units. These units have 3-fold radial symmetry at two vertices and 5-fold radial symmetry at one, with 2-fold radial symmetry at the line opposite of the 5-fold vertex and a 2/5 circular fold wall surrounding the point of the 5-fold vertex. Twenty 3-fold vertices, thirty 2-fold lines, and twelve 5-fold vertices exist per capsid, the latter corresponding to the 12 vertices of the icosahedron.[2]
Typical features of the capsid surface include depressions at each 2-fold axis, elevated protrusions surrounding the 3-fold axes, and raised cylindrical projections made of five beta-barrels[6] surrounded by canyon-like depressions at the 5-fold axes. Each of these cylinders potentially contains an opening to connect the exterior of the capsid to the interior, which mediates entry and exit of the genome. About 20 nucleotides from the 5′-end of the genome may remain exposed outside of the capsid carrying a copy of NS1 bound to the 5′-end, which is a result of how the genome is synthesized and packaged.[2]
Varying sizes of the VP protein are expressed for different parvoviruses, the smaller ones, VP2–5, being expressed at a higher frequency than the large size, VP1. The smaller VPs share a common C-terminus with different N-terminus lengths due to truncation. For VP1, the N-terminus is extended to contain regions important in the replication cycle, and it is incorporated into the capsid, typically 5–10 per capsid, with the common C-terminus responsible for assembling capsids.[1][2]
Each VP monomer contains a core beta-barrel structure called the jelly roll motif of eight strands arranged in two adjacent antiparallel beta sheets, labeled CHEF and BIDG after the individual strands, the latter forming the interior surface of the capsid. Individual beta strands are connected by loops that have varying length, sequence, and conformation, and most of these loops extend toward the exterior surface, giving parvoviruses their unique, rough surface. Related parvoviruses share their surface topologies and VP protein folds to a greater degree than their sequence identities, so the structure of the capsid and capsid protein are useful indicators of phylogeny.[1][2]
Life cycle[edit]
Parvoviruses enter cells by endocytosis, using a variety of cellular receptors to bind to the host cell. In endosomes, many parvoviruses undergo a change in conformation so that the phospholipase A2 (PLA2) domain on the VP1 N-termini are exposed so the virion can penetrate lipid bilayer membranes. Intracellular trafficking of virions varies, but virions ultimately arrive to the nucleus, inside of which the genome is uncoated from the capsid. Based on studies of minute virus of mice (MVM), the genome is ejected from the capsid in a 3′-to-5′ direction from one of the openings in the capsid, leaving the 5′-end of the DNA attached to the capsid.[2]
Parvoviruses lack the ability to induce cells into their DNA replication stage, called S-phase, so they must wait in the nucleus until the host cell enters S-phase on its own. This makes cell populations that divide rapidly, such as fetal cells, an excellent environment for parvoviruses. Adeno-associated viruses (AAV) are dependent on helper viruses, which may be an adenovirus or a herpesvirus, since coinfection alters the cellular environment to allow for replication.[2] In the absence of coinfection, AAV's genome is integrated into the host cell's genome until coinfection occurs.[7] Infected cells that enter S-phase are forced to synthesize viral DNA and cannot leave S-phase. Parvoviruses establish replication foci in the nucleus that grow progressively larger as infection progresses.[8]
Once a cell enters S-phase and the genome is uncoated, a host DNA polymerase uses the 3′-end of the 3′ hairpin as a primer to synthesize a complementary DNA strand for the coding portion of the genome, which is connected to the 5′-end of the 5′ hairpin.[3][7][9] Messenger RNA (mRNA) that encodes NS1 is then transcribed from the genome by the DNA polymerase, capped and polyadenylated, and translated by host ribosomes to synthesize NS1.[2][5][10] If proteins are encoded in multiple co-linear frames, then alternative splicing, suboptimal translation initiation, or leaky scanning may be used to translate different gene products.[2]
Parvoviruses replicate their genome via rolling hairpin replication, a unidirectional, strand displacement form of DNA replication that is initiated by NS1. Replication begins once NS1 binds to and makes a nick in a replication origin site in the duplex DNA molecule at the end of one hairpin. Nicking releases the 3′-end of the nicked strand as a free hydroxyl (-OH) to prime DNA synthesis[2] with NS1 remaining attached to the 5′-end.[7] The nick causes the adjacent hairpin to unfold into a linear, extended form. At the 3′-OH, a replication fork is established using NS1's helicase activity, and the extended telomere is replicated by the DNA polymerase.[10][11] The two telomere strands then refold back in on themselves to their original configurations, which repositions the replication fork to switch templates to the other strand and move in the opposite direction toward the other end of the genome.[12][13]
Parvoviruses vary in whether the termini are similar or the same, called homotelomeric parvoviruses, or different, called heterotelomeric parvoviruses. In general, homotelomeric parvoviruses, such as AAV and B19, replicate both ends of their genome through the aforementioned process, called terminal resolution, and their hairpin sequences are contained within larger (inverted) terminal repeats. Heterotelomeric viruses, such as minute virus of mice (MVM), replicate one end by terminal resolution and the other end via an asymmetric process called junction resolution[2][14] so that the correct orientation of the telomere can be copied.[15]
During asymmetric junction resolution, the duplex extended-form telomeres refold in on themselves into a cruciform shape. A replication origin site on the lower strand of the right arm of the cruciform is nicked by NS1, leading to the lower arm of the cruciform unfolding into its linear extended form. A replication fork established at the nick site moves down the extended lower arm to copy the lower arm's sequence. The two strands of the lower arm then refold to reposition the replication fork to go back toward the other end, displacing the upper strand in the process.[16]
The back and forth, end-to-end pattern of rolling hairpin replication produces a concatemer containing multiple copies of the genome.[2][3] NS1 periodically makes nicks in this molecule and, through a combination of terminal resolution and junction resolution, individual strands of the genome are excised from the concatemer.[9][13] Excised genomes may either be recycled for further rounds of replication or packaged into progeny capsids.[7] Translation of mRNA containing VP proteins leads to the accumulation of capsid proteins in the nucleus that assemble into these empty capsids.[8]
Genomes are encapsidated at one of the capsid's vertices through a portal,[2] potentially the one opposite the portal used to expel the genome.[5] Once complete virions have been constructed, they may be exported from the nucleus to the exterior of the cell before disintegration of the nucleus. Disruption of the host cell environment may also occur later on in the infection. This results in cell lysis via necrosis or apoptosis, which releases virions to the outside of the cell.[2][8]
Evolution[edit]
Parvoviruses are believed to be descended from ssDNA viruses that have a circular genome that forms a loop and which replicate via rolling circle replication, which is similar to rolling hairpin replication. These circular ssDNA viruses encode a replication initiator protein that is related to and possesses many of the same characteristics as the replication initiator protein of parvoviruses, such as the HUH endonuclease domain and the SF3 helicase domain.[17] In contrast to these other replication initiator proteins, NS1 shows only vestigial traces of being able to perform ligation, which is a key part of rolling circle replication.[8] The Bidnaviridae family, which are also linear ssDNA viruses, appear to be descended from a parvovirus that had its genome integrated into the genome of a polinton, a type of DNA transposon related to viruses in the realm Varidnaviria.[17]
Based on phylogenetic analysis of the SF3 helicase, parvoviruses split into two branches early in their evolutionary history, one of which contains viruses assigned to the subfamily Hamaparvovirinae. The other branch split into two sublineages that constitute the other two subfamilies, Densovirinae and Parvovirinae.[18] Parvoviruses in the Hamaparvovirinae lineage are likely all heterotelomeric, Densovirinae are exclusively homotelomeric, and Parvovirinae varies.[2] Telomere sequences have significant complexity and diversity, suggesting that many species have co-opted them to perform additional functions.[7][10] Parvoviruses are also considered to have high rates of genetic mutations and recombinations.[2][9]
Parvoviruses constitute the family Parvoviridae. The family is the sole family in the order Piccovirales, which is the sole order in the class Quintoviricetes. The class Quintoviricetes belongs to the phylum Cossaviricota, which also includes papillomaviruses, polyomaviruses, and bidnaviruses. Cossaviricota is included in the kingdom Shotokuvirae, which is assigned to the realm Monodnaviria. Parvoviridae belongs to Group II: ssDNA viruses in the Baltimore classification system, which groups viruses together based on their manner of mRNA synthesis. Within Parvoviridae, three subfamilies, 26 genera, and 126 species are recognized as of 2020 (-virinae denotes subfamily and -virus denotes genus):[18][19]
- Densovirinae (11 genera, 21 species)
- Aquambidensovirus (3 species)
- Blattambidensovirus (1 species)
- Diciambidensovirus (1 species)
- Hemiambidensovirus (2 species)
- Iteradensovirus (5 species)
- Miniambidensovirus (1 species)
- Muscodensovirus (1 species)
- Pefuambidensovirus (1 species)
- Protoambidensovirus (2 species)
- Scindoambidensovirus (3 species)
- Tetuambidensovirus (1 species)
- Hamaparvovirinae (5 genera, 21 species)
- Brevihamaparvovirus (2 species)
- Chaphamaparvovirus (16 species)
- Hepanhamaparvovirus (1 species)
- Ichthamaparvovirus (1 species)
- Penstylhamaparvovirus (1 species)
- Parvovirinae (10 genera, 84 species)
- Amdoparvovirus (5 species)
- Artiparvovirus (1 species)
- Aveparvovirus (3 species)
- Bocaparvovirus (28 species)
- Copiparvovirus (7 species)
- Dependoparvovirus (11 species)
- Erythroparvovirus (7 species)
- Loriparvovirus (1 species)
- Protoparvovirus (15 species)
- Tetraparvovirus (6 species)
Parvoviruses are assigned to the same species if they share at least 85% of their protein sequence identities. Species are grouped together in a genus based on phylogeny of the NS1 and SF3 helicase domains, as well as similarity of NS1 sequence identity and coverage. If these criteria aren't satisfied, then genera can still be established provided that common ancestry is supported. The three subfamilies are distinguished based on phylogeny of the SF3 helicase domain, which corresponds to host range: viruses in Densovirinae infect invertebrates, viruses in Hamaparvovirinae infect invertebrates and vertebrates, and viruses in Parvovirinae infect vertebrates.[18]
In humans, the most prominent parvoviruses that cause disease are parvovirus B19 and human bocavirus 1. B19 infection is often asymptomatic but can manifest in a variety of ways, including Fifth disease with its characteristic rash in children, persistent anemia in immunocompromised persons and in people who have underlying hemoglobinopathies,[20] transient aplastic crises, hydrops fetalis in pregnant women, and arthropathy. Human bocavirus 1 is a common cause of acute respiratory tract infection, especially in young children, wheezing being a common symptom. Other parvoviruses associated with different diseases in humans include human parvovirus 4 and human bufavirus, though the manner by which these viruses cause disease is unclear.[6]
Carnivore-infecting viruses in the genus Protoparvovirus, in contrast to human parvoviruses, are more life-threatening.[2] Canine parvoviruscauses severe illness in dogs, the most common symptom being hemorrhagic enteritis, with up to a 70% mortality rate in pups but usually less than 1% in adults.[21] Feline parvovirus, a closely related virus,[22] likewise causes severe illness in cats along with panleukopenia.[23][24] In pigs, porcine parvovirus is a major cause of infertility as infection frequently leads to death of the fetus.[25]
Use in medicine[edit]
Adeno-associated viruses have become an important vector for gene therapy aimed at treating genetic diseases, such as those caused by a single mutation. The recombinant AAV (rAAV) contains a viral capsid but lacks a complete viral genome. Instead, the typical nucleic acid packaged into the capsid contains a promoter region, the gene of interest, and a terminator region, all contained within two inverted terminal repeats derived from the viral genome. rAAV essentially acts as a container that can traverse the cell membrane and deliver its nucleic acid cargo to the nucleus.[26][27]
https://en.wikipedia.org/wiki/Parvoviridae
Ichthyosis vulgaris (also known as "autosomal dominant ichthyosis"[1] and "Ichthyosis simplex"[1]) is a skin disorder causing dry, scaly skin. It is the most common form of ichthyosis,[2]:486 affecting around 1 in 250 people.[3] For this reason it is known as common ichthyosis. It is usually an autosomal dominant inherited disease (often associated with filaggrin), although a rare non-heritable version called acquired ichthyosis exists.[4]:560
| Ichthyosis vulgaris | |
|---|---|
 | |
| Ichthyosis vulgaris #1 (top-left) | |
| Specialty | Medical genetics |
https://en.wikipedia.org/wiki/Ichthyosis_vulgaris
Keratosis pilaris (also follicular keratosis, lichen pilaris, or colloquially chicken skin[1]) is a common, autosomal-dominant, genetic condition of the skin's hair follicles characterized by the appearance of possibly itchy, small, gooseflesh-like bumps, with varying degrees of reddening or inflammation.[2] It most often appears on the outer sides of the upper arms (the forearms can also be affected), thighs, face, back, and buttocks;[2] KP can also occur on the hands, and tops of legs, sides, or any body part except glabrous (hairless) skin (like the palms or soles of feet).[3] Often the lesions can appear on the face, which may be mistaken for acne.[4]
The several types of KP have been associated with pregnancy, type 1 diabetes mellitus, obesity, dry skin, allergic diseases (e.g., atopic dermatitis), and rarely cancer.[1] Many rarer types of the disorder are part of inherited genetic syndromes.[1]
The cause of KP is not completely understood. As of 2018, KP is thought to be due to abnormalities in the process of depositing the protein keratin in hair follicles, abnormalities in the hair shaft, or both.[1] KP is usually diagnosed by a medical professional based on the appearance of the skin, but dermoscopy can be used, as well, if the diagnosis is unclear.[1] Variants of the ABCA12 gene have been associated with KP.[5]
| Keratosis pilaris | |
|---|---|
| Other names | Follicular keratosis, lichen pilaris |
 | |
| Condition on a calf | |
| Specialty | Dermatology |
BRAF is a human gene that encodes a protein called B-Raf. The gene is also referred to as proto-oncogene B-Rafand v-Raf murine sarcoma viral oncogene homolog B, while the protein is more formally known as serine/threonine-protein kinase B-Raf.[5][6]
The B-Raf protein is involved in sending signals inside cells which are involved in directing cell growth. In 2002, it was shown to be mutated in some human cancers.[7]
Certain other inherited BRAF mutations cause birth defects.
Drugs that treat cancers driven by BRAF mutations have been developed. Two of these drugs, vemurafenib[8] and dabrafenib are approved by FDA for treatment of late-stage melanoma. Vemurafenib was the first approved drug to come out of fragment-based drug discovery.[9]
https://en.wikipedia.org/wiki/BRAF_(gene)#BRAF_inhibitors_in_the_clinic
News Articles Online
AP News
No evidence that COVID-19 vaccine results in sterilization (Dupuy, 2020)
CLAIM: The head of research at Pfizer says the COVID-19 vaccine causes female sterilization because it contains a spike protein known as syncytin-1.
https://apnews.com/article/fact-checking-9856420671
The role of syncytins in human reproduction and reproductive organ cancers
in Reproduction
Authors: Bikem Soygur 1 and Leyla Sati 1
Human life begins with sperm and oocyte fusion. After fertilization, various fusion events occur during human embryogenesis and morphogenesis. For example, the fusion of trophoblastic cells constitutes a key process for normal placental development. Fusion in the placenta is facilitated by syncytin 1 and syncytin 2. These syncytins arose from retroviral sequences that entered the primate genome 25 million and more than 40 million years ago respectively. About 8% of the human genome consists of similar human endogenous retroviral (HERVs) sequences. Many are inactive because of mutations or deletions. However, the role of the few that remain transcriptionally active has not been fully elucidated. Syncytin proteins maintain cell–cell fusogenic activity based on env gene-mediated viral cell entry. In this review, we summarize how syncytins and their receptors are involved in fusion events during human reproduction. The significance of syncytins in tumorigenesis is also discussed.
Human life begins with sperm and oocyte fusion. After fertilization, various fusion events occur during human embryogenesis and morphogenesis. For example, the fusion of trophoblastic cells constitutes a key process for normal placental development. Fusion in the placenta is facilitated by syncytin 1 and syncytin 2. These syncytins arose from retroviral sequences that entered the primate genome 25 million and more than 40 million years ago respectively. About 8% of the human genome consists of similar human endogenous retroviral (HERVs) sequences. Many are inactive because of mutations or deletions. However, the role of the few that remain transcriptionally active has not been fully elucidated. Syncytin proteins maintain cell–cell fusogenic activity based on env gene-mediated viral cell entry. In this review, we summarize how syncytins and their receptors are involved in fusion events during human reproduction. The significance of syncytins in tumorigenesis is also discussed.
Syncytin 1 (HERV-W; ERVW-1) and human placentation
The role of retroviral proteins, especially syncytins, in trophoblastic fusion process and placental morphogenesis was hypothesized about 15 years ago (Mi et al. 2000, Blaise et al. 2003). Mi et al. (2000) first identified syncytin 1 in the syncytiotrophoblast layer of human placental villi (Mi et al. 2000). They showed that when syncytin 1 is transfected into COS cells (CV-1 in origin and carrying the SV40 genetic material), syncytia formed consisting of many aggregated nuclei surrounded by an extended cytoplasm (Mi et al. 2000). When BeWo cells (trophoblast-derived choriocarcinoma cell line) are induced by forskolin to fuse and form syncytiotrophoblast-like cells, a five-fold increase in BeWo cell fusion is correlated with increased syncytin (ERVW-1) transcription (Mi et al. 2000). Thus, a role of syncytin 1 in placental cytotrophoblast fusion and its fusogenic properties in vitro is demonstrated (Mi et al.2000). Following the discovery of syncytin 1, Blond et al. (2000) showed that transfection of different cell lines with syncytin 1 results in syncytia formation via the interaction of syncytin 1 and its receptor, SLC1A5 (Blond et al. 2000). SLC1A5 expression is reported in villous (Soygur et al. 2016) and extravillous trophoblast (Malassine et al. 2005).
After identification of the fundamental fusion role of syncytin 1 and upstream components in this signaling pathway, such as CD9 and cAMP/PKA (Muroi et al. 2009), many reports have shown the presence of syncytin 1 in the basal membrane of syncytiotrophoblast (Lee et al. 2001), cytotrophoblast (Blond et al. 2000, Smallwood et al. 2003, Muir et al. 2006, Soygur et al. 2016), and some stromal cells in the core of placental villi (Holder et al. 2012, Soygur et al.2016). It is of interest to note that the presence of syncytin 1 in the stromal core of villi may further indicate some additional non-fusogenic functions of syncytin 1. The presence of syncytin 1 in the apical microvillous membrane of villous trophoblast is also reported by our laboratory (Soygur et al. 2016). Malassine et al. (2005) showed syncytin 1 expression in all cell types of the extravillous phenotype lineage (Malassine et al. 2005). Cytotrophoblast in the tips of villi can differentiate into another type of trophoblast called the extravillous trophoblast. However, extravillous trophoblast cells are anchored and invade into the deeper layers of the decidua and maternal vascular bed (Cartwright et al. 2010). As cell–cell fusion of extravillous trophoblast does not occur at the maternal–fetal interface, syncytin 1 expression in extravillous trophoblast arouses great interest. Glial cells missing 1 (GCM1) is an important placental transcription factor as chorionic trophoblast cells in Gcm1-deficient placentas do not fuse to form syncytiotrophoblast (Anson-Cartwright et al. 2000). Wang et al. (2012) identified the GCM1 target gene, HtrA4 (high-temperature requirement protein A4), and reported that HtrA4 protein mediates placental JAR (choriocarcinoma cell line) and BeWo cell invasion by cleaving the extracellular matrix (ECM) protein fibronectin. More importantly, their study also demonstrated that HtrA4 suppresses the cell–cell fusion mediated by syncytin 1 in transfected human embryonic kidney 293T cell line for the first time. Binding of HtrA4 PZD domain to the SU subunit of syncytin results in decreased syncytin 1 expression on the cell surface. While HtrA4 decreases syncytin 1-mediated cell fusion, it also supports the invasion of JAR and BeWo cells in vitro. Overall, the results indicated the importance of HtrA4 and syncytin 1 in extravillous trophoblast differentiation by preventing cell fusion and promoting invasion in extravillous trophoblast (Wang et al. 2012).
On the other hand, Huang et al. (2013) investigated the role of syncytin 1 in trophoblast proliferation (Huang et al.2013). In this study, syncytin 1 knockdown by siRNA significantly inhibited BeWo cell growth and DNA synthesis in vitro. Analysis of G1/S cell cycle checkpoint regulators in syncytin 1 knockdown BeWo cells showed that there were decreased CDK4, E2F1, PCNA and c-Myc levels in contrast to increased p15 protein level after siRNA transfection. At 72-h post-transfection in syncytin 1 knockdown BeWo cells compared with control groups, there was an increased percentage of cells in G1 phase and a decreased percentage in S and G2/M phases. These researchers therefore showed that syncytin 1 knockdown causes cell cycle arrest at the G1 phase (Huang et al. 2013). Because mononucleated cytotrophoblastic cells leave the cell cycle to differentiate into multinucleated syncytiotrophoblasts, they no longer have the ability to proliferate (Benirschke & Kaufmann 2000). In case of insufficient syncytin 1 protein, cell cycle arrest may occur in cytotrophoblasts. The inadequate proliferation of cytotrophoblast and the absence of continuous fusion with syncytiotrophoblast may result in impairment of the syncytiotrophoblast layer. Therefore, one can speculate that syncytin 1 is possibly not only involved in the fusion of cytotrophoblast but also the proliferation of the cytotrophoblast via cell cycle. These independent properties of syncytin 1 (fusogenic and non-fusogenic) can maintain a balance between the ‘cytotrophoblast pool’ and the syncytiotrophoblast layer during human placental development.
Tolosa et al. (2012) showed a possible immune regulatory function of syncytin 1 in vitro (Tolosa et al. 2012). It is known that Th1 cytokines (e.g., TNF-α, IFN-ɣ and IL-2) have harmful effects on the fetus and are downregulated during pregnancy (Raghupathy 1997). Tolosa et al. (2012) reported that lipopolysaccharide/phytohemagglutinin (LPS/PHA)-stimulated Th1 cytokine responses (TNF-α and IFN-ɣ) and chemokine CXCL10 are inhibited by a syncytin 1 recombinant ectodomain in a human blood culture system. Moreover, CRH (corticotropin-releasing hormone) treatment of BeWo cells increases secreted exosomal syncytin 1 protein expression but not cellular syncytin 1. These results suggest that the presence of syncytin 1 in placental exosomes might provide a mechanism for syncytin 1 to reach and interact with target cells of the maternal immune system during pregnancy (Tolosa et al. 2012).
Further studies are carried out to understand the potential roles of syncytin 1 in placental pathologies such as preeclampsia (PE) (Lee et al. 2001, Chen et al. 2006, Vargas et al. 2011, Holder et al. 2012), intrauterine growth restriction (IUGR), and gestational diabetes mellitus (GDM) (Soygur et al. 2016). PE is a pregnancy-related disorder that affects approximately 2–7% of all pregnancies (Acien et al. 1990). In PE pregnancies, poor replacement of spiral artery wall by endovascular trophoblasts and insufficient placental circulation result in oxidative stress, hypoxia and endothelial dysfunction (Benirschke & Kaufmann 2000). PE is diagnosed based on arterial hypertension and proteinuria in pregnancy (Wilson et al. 2003). Many reports have shown decreased expression and aberrant localization of syncytin 1 in PE placentas compared with healthy controls (Lee et al. 2001, Chen et al.2006, Langbein et al. 2008, Vargas et al. 2014, Zhuang et al. 2014). Chiang et al. (2009) showed decreased levels of GCM1, syncytin 1 and placental growth factor, which are all crucial for syncytiotrophoblast formation and placental vasculogenesis in PE placentas. While hypoxia enables activation of glycogen synthase kinase 3 beta (GSK-3β) in PE placenta, the PI3K–Akt pathway is inactivated under hypoxic condition in PE placental cells. Activated GSK-3β phosphorylates GCM1, promotes its ubiquitination, and is then degraded by the SCFFBW2 E3 ligase. As a result of disruption of the GCM1 transcription network, its target genes, syncytin 1 and placental growth factor, are decreased in a parallel manner (Chiang et al. 2009). Studies have also indicated a relationship between syncytin 1 and apoptosis in PE placentas (Ishihara et al. 2002, Huang et al. 2014b). Syncytin 1 knockdown in BeWo cells results in increased apoptosis in this carcinoma cell line of trophoblastic origin. Surprisingly, apoptosis in BeWo cells is mediated by apoptosis-inducing factor (AIF), which is independent of caspase. Decreased syncytin 1, increased AIF and increased calpain1 protein levels in apoptotic cells of human PE placentas have also been shown (Huang et al. 2014b). Thus, changes in cell cycle progression and apoptosis caused by altered syncytin expression may cause abnormalities in PE placentas.
The role of syncytin 1 in intrauterine growth-restricted (IUGR) placentas has also been investigated. The chorionic villi surface areas are reduced compared with age-related controls, and a smaller interface between maternal and fetal tissues is observed in IUGR placentas (Biswas et al. 2008). Moreover, an abnormal cellular development of trophoblast and increased trophoblast apoptosis are also seen (Ishihara et al. 2002). Ruebner et al. (2010) showed decreased syncytin 1 levels in IUGR placentas, which may contribute to placental dysfunction in IUGR (Ruebner et al. 2010). Although deregulation of syncytin 1 in PE placental pathology has been comprehensively studied, the role of syncytin 1 in fetal growth retardation is less certain. Thus, further functional studies are needed to highlight the regulatory mechanisms of syncytin 1 in IUGR placentas.
A recent report has identified the first host cell-encoded inhibitor protein, termed suppressyn, for mammalian cell fusion. Like syncytin, suppressyn is HERV-derived, placenta-specific and conserved during evolution. Suppressyn protein inhibits syncytin 1-induced cell fusion by binding with the syncytin 1 receptor, SLC1A5, but does not affect syncytin 2-mediated syncytialization (Sugimoto et al. 2013). Unfortunately, the role and regulatory mechanisms of syncytin 1 in different placental pathologies are still not known clearly. Thus, identification and involvement of suppressyn in syncytin 1-mediated cell fusion during placentation might provide a useful approach to better understand the underlying molecular mechanisms in placental pathologies.
Syncytin 2 (HERV-FRD; ERVFRD-1) and human placentation
Syncytin 2 was initially characterized in human placenta by screening human sequence databases for endogenous envelope retroviral elements (Blaise et al. 2003). When 16 candidate ENV retroviral genes were cloned in a eukaryotic expression vector and fusogenic activity was determined in transfected mammalian cells, syncytin 2 (ERVFRD-1) was discovered. Esnault et al. (2008) showed that syncytin 2 interacts with a different receptor (MFSD2) than syncytin 1 (Esnault et al. 2008). Further studies analyzed the amino acid sequence of syncytin 2 and demonstrated an immunosuppressive domain (Mangeney et al. 2007). This domain may protect the fetus against the maternal immune system (Rawn & Cross 2008, Lavialle et al. 2013). Data reported by Mangeney et al. (2007), using an in vivo tumor rejection assay, also supports the idea that syncytin 2, but not syncytin 1, has immunosuppressive activity. Tumorigenicity potential was assessed after syncytin 1- or syncytin 2-transduced MCA205 cells were engrafted to mice. Even though syncytin 2-transduced MCA205 cells formed large long-lasting tumors, syncytin 1-transduced MCA205 cells formed small tumors that were rapidly eliminated (Mangeney et al.2007).
https://rep.bioscientifica.com/view/journals/rep/152/5/R167.xml
One such post (here) reads: “The mRNA vacks targets a protein called Syncitin-1 which is a protein contained in the CV spike protein complex.
“The PROBLEM is, there is no mechanism to turn off or STOP the destruction of Syncitin-1 which is also found in the BRAIN and female reproductive system. Therefore, Syncitin-1 will continue to be scavenged in the body. Without Syncitin-1 no placenta can form. The brain manifestations of no Syncitin-1 are MS, Parkinsons, Schizophrenia, Psychopathy and I posit; any neurodegenerative condition.” [sic]
The Pfizer/BioNTech and Moderna COVID-19 vaccines work by inserting mRNA which tells human cells to produce a sequence of amino acids that forms a protein like the “spike protein” found on the SARS-CoV-2 virus which causes COVID-19. The mRNA is then destroyed (here).
https://www.reuters.com/article/uk-factcheck-syncytin/fact-check-available-mrna-vaccines-do-not-target-syncytin-1-a-protein-vital-to-successful-pregnancies-idUSKBN2A42S7
Coronavirus Proteins
New Spike Mutant Proteins – Alpha, Beta, Gamma, Delta Variants
Company Information
https://www.rndsystems.com/products/proteins-coronavirus-research?gclid=Cj0KCQjw-NaJBhDsARIsAAja6dNMuCvZDQSMl7inMLV3HkOSJLqrpi9aA8AlGFBYADsDHoZU33rQXC8aAgKEEALw_wcB
REVIEW article
Impact of COVID-19 on Mitochondrial-Based Immunity in Aging and Age-Related Diseases
https://www.frontiersin.org/articles/10.3389/fnagi.2020.614650/full
The role ofOncotarget. 2017 Nov 10; 8(56): 95945–95964.
Published online 2017 Oct 7. doi: 10.18632/oncotarget.21606
PMCID: PMC5707072
PMID: 29221178
Cytotoxic stress induces transfer of mitochondria-associated human endogenous retroviral RNA and proteins between cancer cells
David Díaz-Carballo,1 Jacqueline Klein,1 Ali H. Acikelli,1 Camilla Wilk,1 Sahitya Saka,1 Holger Jastrow,2Gunther Wennemuth,2 Phillip Dammann,3 Urs Giger-Pabst,4 Veria Khosrawipour,4 Joachim Rassow,5 Mikalai Nienen,6and Dirk Strumberg1
About 8 % of the human genome consists of human endogenous retroviruses (HERVs), which are relicts of ancient exogenous retroviral infections incurred during evolution. Although the majority of HERVs have functional gene defects or epigenetic modifications, many of them are still able to produce retroviral proteins that have been proposed to be involved in cellular transformation and cancer development.
The role ofOncotarget. 2017 Nov 10; 8(56): 95945–95964.
Published online 2017 Oct 7. doi: 10.18632/oncotarget.21606
PMCID: PMC5707072
PMID: 29221178
Cytotoxic stress induces transfer of mitochondria-associated human endogenous retroviral RNA and proteins between cancer cells
David Díaz-Carballo,1 Jacqueline Klein,1 Ali H. Acikelli,1 Camilla Wilk,1 Sahitya Saka,1 Holger Jastrow,2Gunther Wennemuth,2 Phillip Dammann,3 Urs Giger-Pabst,4 Veria Khosrawipour,4 Joachim Rassow,5 Mikalai Nienen,6and Dirk Strumberg1
We found that, in chemo-resistant U87RETO glioblastoma cells, cytotoxic stress induced by etoposide promotes accumulation and large-scale fission of mitochondria, associated with the detection of HERV-WE1 (syncytin-1) and HERV-FRD1 (syncytin-2) in these organelles. In addition, mitochondrial preparations also contained the corresponding receptors, i.e. ASCT2 and MFSD2. We clearly demonstrated that mitochondria associated with HERV-proteins were shuttled between adjacent cancer cells not only via tunneling tubes, but also by direct cellular uptake across the cell membrane. Furthermore, anti-syncytin-1 and anti-syncytin-2 antibodies were able to specifically block this direct cellular uptake of mitochondria even more than antibodies targeting the cognate receptors.
Here, we suggest that the association of mitochondria with syncytin-1/syncytin-2 together with their respective receptors could represent a novel mechanism of cell-to-cell transfer. In chemotherapy-refractory cancer cells, this might open up attractive avenues to novel mitochondria-targeting therapies.
Keywords: human endogenous retroviruses (HERVs), mitochondria, intercellular mitochondrial transfer, syncytin-1/2 receptors
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5707072/
https://pubmed.ncbi.nlm.nih.gov/29221178/
https://academic.oup.com/molehr/article/21/2/206/973472
https://academic.oup.com/jhered/article/97/2/100/2187562
Upregulation of syncytin-1 promotes invasion and metastasis by activating epithelial-mesenchymal transition-related pathway in endometrial carcinoma
Changmin Liu,1,* Jiqin Xu,2,* Feifei Wen,3 Fangfang Yang,1 Xiaoming Li,4 Dianzhong Geng,1 Lei Li,1 Jiming Chen,5 Jing Zheng6
2018
https://www.dovepress.com/upregulation-of-syncytin-1-promotes-invasion-and-metastasis-by-activat-peer-reviewed-fulltext-article-OTT
Mitochondria, pluripotent/monocyte/undifferentiated/red blood cell/etc., tissues differentiated cell distribution, extrachromosomal dna, dna repositories, recombination/etc., other code/etc.
brain reproduc syncytin
herv sars covid mitochondria hiv aids
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5707072/
Authors: Bikem Soygur 1 and Leyla Sati 1
https://rep.bioscientifica.com/view/journals/rep/152/5/R167.xml
Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA (in this context called complementary DNA or cDNA) and amplification of specific DNA targets using polymerase chain reaction (PCR).[1] It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.
The close association between RT-PCR and qPCR has led to metonymic use of the term qPCR to mean RT-PCR. Such use may be confusing,[2] as RT-PCR can be used without qPCR, for example to enable molecular cloning, sequencing or simple detection of RNA. Conversely, qPCR may be used without RT-PCR, for example to quantify the copy number of a specific piece of DNA.
https://en.wikipedia.org/wiki/Reverse_transcription_polymerase_chain_reaction
https://www.researchgate.net/post/How-can-i-synthesize-cDNA-of-miRNA
https://www.thermofisher.com/order/catalog/product/4366596
In genetics, complementary DNA (cDNA) is DNA synthesized from a single-stranded RNA (e.g., messenger RNA (mRNA) or microRNA (miRNA)) template in a reaction catalyzed by the enzyme reverse transcriptase. cDNA is often used to clone eukaryoticgenes in prokaryotes. When scientists want to express a specific protein in a cell that does not normally express that protein (i.e., heterologous expression), they will transfer the cDNA that codes for the protein to the recipient cell. In molecular biology, cDNA is also generated to analyze transcriptomic profiles in bulk tissue, single cells, or single nuclei in assays such as microarrays and RNA-seq.
cDNA is also produced naturally by retroviruses (such as HIV-1, HIV-2, simian immunodeficiency virus, etc.) and then integrated into the host's genome, where it creates a provirus.[1]
The term cDNA is also used, typically in a bioinformatics context, to refer to an mRNA transcript's sequence, expressed as DNA bases (deoxy-GCAT) rather than RNA bases (GCAU).
https://en.wikipedia.org/wiki/Complementary_DNA
In genetics, complementation occurs when two strains of an organism with different homozygous recessive mutations that produce the same mutant phenotype (for example, a change in wing structure in flies) have offspring that express the wild-type phenotype when mated or crossed. Complementation will ordinarily occur if the mutations are in different genes (intergenic complementation). Complementation may also occur if the two mutations are at different sites within the same gene (intragenic complementation), but this effect is usually weaker than that of intergenic complementation. In the case where the mutations are in different genes, each strain's genome supplies the wild-type allele to "complement" the mutated allele of the other strain's genome. Since the mutations are recessive, the offspring will display the wild-type phenotype. A complementation test (sometimes called a "cis-trans" test) can be used to test whether the mutations in two strains are in different genes. Complementation ordinarily will occur more weakly or not at all if the mutations are in the same gene. The convenience and essence of this test is that the mutations that produce a phenotype can be assigned to different genes without the exact knowledge of what the gene product is doing on a molecular level. The complementation test was developed by Americangeneticist Edward B. Lewis.
If the combination of two genomes containing different recessive mutations yields a mutant phenotype, then there are three possibilities:
- Mutations occur in the same gene.
- One mutation affects the expression of the other.
- One mutation may result in an inhibitory product.
Deoxyribonucleic acid (/diːˈɒksɪˌraɪboʊnjuːˌkliːɪk, -ˌkleɪ-/ (![]() listen);[1] DNA) is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.
listen);[1] DNA) is a molecule composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.
The two DNA strands are known as polynucleotides as they are composed of simpler monomeric units called nucleotides.[2][3] Each nucleotide is composed of one of four nitrogen-containing nucleobases (cytosine [C], guanine [G], adenine [A] or thymine [T]), a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds (known as the phospho-diester linkage) between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are bound together, according to base pairing rules (A with T and C with G), with hydrogen bonds to make double-stranded DNA. The complementary nitrogenous bases are divided into two groups, pyrimidines and purines. In DNA, the pyrimidines are thymine and cytosine; the purines are adenine and guanine.
Both strands of double-stranded DNA store the same biological information. This information is replicated as and when the two strands separate. A large part of DNA (more than 98% for humans) is non-coding, meaning that these sections do not serve as patterns for protein sequences. The two strands of DNA run in opposite directions to each other and are thus antiparallel. Attached to each sugar is one of four types of nucleobases (or bases). It is the sequence of these four nucleobases along the backbone that encodes genetic information. RNA strands are created using DNA strands as a template in a process called transcription, where DNA bases are exchanged for their corresponding bases except in the case of thymine (T), for which RNA substitutes uracil (U).[4] Under the genetic code, these RNA strands specify the sequence of amino acids within proteins in a process called translation.
Within eukaryotic cells, DNA is organized into long structures called chromosomes. Before typical cell division, these chromosomes are duplicated in the process of DNA replication, providing a complete set of chromosomes for each daughter cell. Eukaryotic organisms (animals, plants, fungi and protists) store most of their DNA inside the cell nucleus as nuclear DNA, and some in the mitochondria as mitochondrial DNA or in chloroplasts as chloroplast DNA.[5] In contrast, prokaryotes (bacteria and archaea) store their DNA only in the cytoplasm, in circular chromosomes. Within eukaryotic chromosomes, chromatin proteins, such as histones, compact and organize DNA. These compacting structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
https://en.wikipedia.org/wiki/DNA
A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.
Most prokaryote chromosomes contain a circular DNA molecule – there are no free ends to the DNA. Free ends would otherwise create significant challenges to cells with respect to DNA replication and stability. Cells that do contain chromosomes with DNA ends, or telomeres (most eukaryotes), have acquired elaborate mechanisms to overcome these challenges. However, a circular chromosome can provide other challenges for cells. After replication, the two progeny circular chromosomes can sometimes remain interlinked or tangled, and they must be resolved so that each cell inherits one complete copy of the chromosome during cell division.
The circular bacteria chromosome replication is best understood in the well-studied bacteria Escherichia coli and Bacillus subtilis. Chromosome replication proceeds in three major stages: initiation, elongation and termination. The initiation stage starts with the ordered assembly of "initiator" proteins at the origin region of the chromosome, called oriC. These assembly stages are regulated to ensure that chromosome replication occurs only once in each cell cycle. During the elongation phase of replication, the enzymes that were assembled at oriC during initiation proceed along each arm ("replichore") of the chromosome, in opposite directions away from the oriC, replicating the DNA to create two identical copies. This process is known as bidirectional replication. The entire assembly of molecules involved in DNA replication on each arm is called a "replisome." At the forefront of the replisome is a DNA helicase that unwinds the two strands of DNA, creating a moving "replication fork". The two unwound single strands of DNA serve as templates for DNA polymerase, which moves with the helicase (together with other proteins) to synthesise a complementary copy of each strand. In this way, two identical copies of the original DNA are created. Eventually, the two replication forks moving around the circular chromosome meet in a specific zone of the chromosome, approximately opposite oriC, called the terminus region. The elongation enzymes then disassemble, and the two "daughter" chromosomes are resolved before cell division is completed.
https://en.wikipedia.org/wiki/Circular_chromosome
Plasmid, Transcriptase, Reverse Transcriptase, polymerase, primase, gyrase, helicase (enzyme, -ase)
Redox
anneal re-anneal - https://en.wikipedia.org/wiki/Annealing_(materials_science)
transcription translation replication
Nucleic acid thermodynamics is the study of how temperature affects the nucleic acid structure of double-stranded DNA (dsDNA). The melting temperature (Tm) is defined as the temperature at which half of the DNA strands are in the random coil or single-stranded (ssDNA) state. Tm depends on the length of the DNA molecule and its specific nucleotide sequence. DNA, when in a state where its two strands are dissociated (i.e., the dsDNA molecule exists as two independent strands), is referred to as having been denatured by the high temperature.
Concepts[edit]
Hybridization[edit]
Hybridization is the process of establishing a non-covalent, sequence-specific interaction between two or more complementary strands of nucleic acids into a single complex, which in the case of two strands is referred to as a duplex. Oligonucleotides, DNA, or RNA will bind to their complement under normal conditions, so two perfectly complementary strands will bind to each other readily. In order to reduce the diversity and obtain the most energetically preferred complexes, a technique called annealing is used in laboratory practice. However, due to the different molecular geometries of the nucleotides, a single inconsistency between the two strands will make binding between them less energetically favorable. Measuring the effects of base incompatibility by quantifying the temperature at which two strands anneal can provide information as to the similarity in base sequence between the two strands being annealed. The complexes may be dissociated by thermal denaturation, also referred to as melting. In the absence of external negative factors, the processes of hybridization and melting may be repeated in succession indefinitely, which lays the ground for polymerase chain reaction. Most commonly, the pairs of nucleic bases A=T and G≡C are formed, of which the latter is more stable.
Denaturation[edit]
DNA denaturation, also called DNA melting, is the process by which double-stranded deoxyribonucleic acid unwinds and separates into single-stranded strands through the breaking of hydrophobic stacking attractions between the bases. See Hydrophobic effect. Both terms are used to refer to the process as it occurs when a mixture is heated, although "denaturation" can also refer to the separation of DNA strands induced by chemicals like formamide or urea.[1]
The process of DNA denaturation can be used to analyze some aspects of DNA. Because cytosine / guanine base-pairing is generally stronger than adenine / thymine base-pairing, the amount of cytosine and guanine in a genome (called the "GC content") can be estimated by measuring the temperature at which the genomic DNA melts.[2] Higher temperatures are associated with high GC content.
DNA denaturation can also be used to detect sequence differences between two different DNA sequences. DNA is heated and denatured into single-stranded state, and the mixture is cooled to allow strands to rehybridize. Hybrid molecules are formed between similar sequences and any differences between those sequences will result in a disruption of the base-pairing. On a genomic scale, the method has been used by researchers to estimate the genetic distance between two species, a process known as DNA-DNA hybridization.[3] In the context of a single isolated region of DNA, denaturing gradient gels and temperature gradient gels can be used to detect the presence of small mismatches between two sequences, a process known as temperature gradient gel electrophoresis.[4][5]
Methods of DNA analysis based on melting temperature have the disadvantage of being proxies for studying the underlying sequence; DNA sequencing is generally considered a more accurate method.
The process of DNA melting is also used in molecular biology techniques, notably in the polymerase chain reaction. Although the temperature of DNA melting is not diagnostic in the technique, methods for estimating Tm are important for determining the appropriate temperatures to use in a protocol. DNA melting temperatures can also be used as a proxy for equalizing the hybridization strengths of a set of molecules, e.g. the oligonucleotide probes of DNA microarrays.
Annealing[edit]
Annealing, in genetics, means for complementary sequences of single-stranded DNA or RNA to pair by hydrogen bonds to form a double-stranded polynucleotide. Before annealing can occur, one of the strands may need to be phosphorylated by an enzyme such as kinase to allow proper hydrogen bonding to occur. The term annealing is often used to describe the binding of a DNA probe, or the binding of a primer to a DNA strand during a polymerase chain reaction. The term is also often used to describe the reformation (renaturation) of reverse-complementary strands that were separated by heat (thermally denatured). Proteins such as RAD52 can help DNA anneal. DNA strand annealing is a key step in pathways of homologous recombination. In particular, during meiosis, synthesis-dependent strand annealing is a major pathway of homologous recombination.
Stacking[edit]
| Step | Melting ΔG°37 (Kcal/mol) |
|---|---|
| T A | -0.12 |
| T G or C A | -0.78 |
| C G | -1.44 |
| A G or C T | -1.29 |
| A A or T T | -1.04 |
| A T | -1.27 |
| G A or T C | -1.66 |
| C C or G G | -1.97 |
| A C or G T | -2.04 |
| G C | -2.70 |
Stacking is the stabilizing interaction between the flat surfaces of adjacent bases. Stacking can happen with any face of the base, that is 5'-5', 3'-3', and vice versa.[7]
Stacking in "free" nucleic acid molecules is mainly contributed by intermolecular force, specifically electrostatic attraction among aromatic rings, a process also known as pi stacking. For biological systems with water as a solvent, hydrophobic effect contributes and helps in formation of a helix.[8] Stacking is the main stabilizing factor in the DNA double helix.[9]
Contribution of stacking to the free energy of the molecule can be experimentally estimated by observing the bent-stacked equilibrium in nicked DNA. Such stabilization is dependent on the sequence.[6] The extent of the stabilization varies with salt concentrations and temperature.[9]
See also[edit]
- Melting point
- Primer (molecular biology) for calculations of Tm
- Base pair
- Complementary DNA
- Western blot
External links[edit]
| Library resources about Nucleic acid hybridization |
- Tm calculations in OligoAnalyzer – Integrated DNA Technologies
- DNA thermodynamics calculations – Tm, melting profile, mismatches, free energy calculations
- Tm calculation – by bioPHP.org.
- https://web.archive.org/web/20080516194508/http://www.promega.com/biomath/calc11.htm#disc
- Invitrogen Tm calculation
- AnnHyb Open Source software for Tm calculation using the Nearest-neighbour method
- Sigma-aldrich technical notes
- Primer3 calculation
- "Discovery of the Hybrid Helix and the First DNA-RNA Hybridization" by Alexander Rich
- uMelt: Melting Curve Prediction
- Tm Tool
- Nearest Neighbor Database: Provides a description of RNA-RNA interaction nearest neighbor parameters and examples of their use.
Annealing may refer to:
- Annealing (biology), in genetics
- Annealing (glass), heating a piece of glass to remove stress
- Annealing (materials science), a heat treatment that alters the microstructure of a material
- Quantum annealing, a method for solving combinatorial optimisation problems and ground states of glassy systems
- Simulated annealing, a numerical optimization technique
Carnivore protoparvovirus 1 (CPPV 1) is a species of parvovirus that infects carnivorans. It causes a highly contagious disease in both dogs and cats. The disease is generally divided into two major genogroups: CPV-1 containing the classical feline panleukopenia virus (FPLV), and CPV-2 containing the canine parvovirus (CPV) which appeared in the 1970s.[2]
FPLV is known to infect all wild and domestic members of the felid (cat) family worldwide.[3] It is a highly contagious, severe infection that causes gastrointestinal, immune system, and nervous system disease. Its primary effect is to decrease the number of white blood cells, causing the disease known as feline panleukopenia.
It was once thought that only CPV-1 or FPLV infects cats.[4] However, it has been confirmed that a feline panleukopenia illness can be caused by CPV 2a, 2b, and 2c.[5][6]
FPLV is commonly referred to as:
- feline infectious enteritis virus (FIE)[3]
- feline parvovirus (FPV or FP or "feline parvo")[7]
- feline parvoviral enteritis[3]
It is sometimes confusingly referred to as "cat plague" and "feline distemper".[8]
In addition to members of the felid family, it can also affect other carnivorans (e.g. raccoon, mink).[3]
| Carnivore protoparvovirus 1 | |
|---|---|
 | |
| Electron micrograph of canine parvovirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Quintoviricetes |
| Order: | Piccovirales |
| Family: | Parvoviridae |
| Genus: | Protoparvovirus |
| Species: | Carnivore protoparvovirus 1 |
| Member virus[1] | |
| |
https://en.wikipedia.org/wiki/Carnivore_protoparvovirus_1
Canine parvovirus (also referred to as CPV, CPV2, or parvo) is a contagious virus mainly affecting dogs. CPV is highly contagious and is spread from dog to dog by direct or indirect contact with their feces. Vaccines can prevent this infection, but mortality can reach 91% in untreated cases. Treatment often involves veterinary hospitalization. Canine parvovirus may infect other mammals including foxes, wolves, cats, and skunks.[1] Felines are susceptible to panleukopenia, a different strain of parvovirus.[2]
| Canine parvovirus | |
|---|---|
 | |
| Electron micrograph of canine parvovirus | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Shotokuvirae |
| Phylum: | Cossaviricota |
| Class: | Quintoviricetes |
| Order: | Piccovirales |
| Family: | Parvoviridae |
| Genus: | Protoparvovirus |
| Species: | |
| Virus: | Canine parvovirus |
https://en.wikipedia.org/wiki/Canine_parvovirus
Hemoglobinopathy is the medical term for a group of inherited blood disorders and diseases that primarily affect red blood cells.[1] They are single-gene disorders and, in most cases, they are inherited as autosomal co-dominant traits.[2]
There are two main groups: abnormal structural hemoglobin variants caused by mutations in the hemoglobin genes, and the thalassemias, which are caused by an underproduction of otherwise normal hemoglobin molecules. The main structural hemoglobin variants are HbS, HbE and HbC. The main types of thalassemia are alpha-thalassemia and beta thalassemia.[3]
The two conditions may overlap because some conditions which cause abnormalities in hemoglobin proteins also affect their production. Some hemoglobin variants do not cause pathology or anemia, and thus are often not classed as hemoglobinopathies.[4][5]
| Hemoglobinopathy | |
|---|---|
| Other names | Hemoglobinopathies |
 | |
| Red blood cells from a person with sickle cell trait | |
| Specialty | Hematology |
Erythema infectiosum, fifth disease, or slapped cheek syndrome[3] is one of several possible manifestations of infection by parvovirus B19.[4] Fifth disease typically presents as a rash and is more common in children. While parvovirus B19 can affect humans of all ages, only two out of ten individuals will present with physical symptoms.[5]
The name "fifth disease" comes from its place on the standard list of rash-causing childhood diseases, which also includes measles (first), scarlet fever (second), rubella (third), Dukes' disease (fourth, but is no longer widely accepted as distinct from scarlet fever), Erythema infectiosum parvovirus (fifth) and roseola (sixth).
| Erythema infectiosum | |
|---|---|
| Other names | Fifth disease, Slapped cheek syndrome, slapcheek, slap face, slapped face[1][2] |
 | |
| 16-month-old with erythema infectiosum | |
| Specialty | Infectious disease |
| Symptoms | Red rash, especially on cheeks |
| Causes | Human parvovirus |
| Gestational | |
|---|---|
| During birth | |
| Late pregnancy | |
| By breastfeeding | |

- File:Portrait of Gerard de Lairesse MET rl1975.1.140.R.jpg
- Created: 1665–67


The popularity of the Panacea is reflected in publications of its time. For example, the product was mentioned in the song The Connecticut Pedlar (c. 1851), where the pedlar's list of offerings includes "Swaim's panacea and Jonses's drops too."[23] And in an 1849 letter to the Southern Literary Messenger, Edgar Allan Poe defended the poetry of Bayard Taylor against critics "who possess little other ability than that which assures temporary success to them in common with Swaim's Panacea or Morrison's Pills." Abolitionist William Lloyd Garrison mentions "taking my third bottle of Swaim's Panacea" for scrofula in an 1836 letter.[24]
One of the most comprehensive accounts of the story of Swaim's Panacea is in the 1961 book The Toadstool Millionaires: A Social History of Patent Medicines in America before Federal Regulation by James Harvey Young.[16]
See also[edit]
| Tabes dorsalis | |
|---|---|
| Other names | Syphilitic myelopathy |
 | |
| Axial section of the spinal cord showing syphilitic destruction (whitened area, upper center) of the posterior columns which carry sensory information from the body to the brain | |
| Specialty | Neurology |
General paresis, also known as general paralysis of the insane (GPI) or paralytic dementia, is a severe neuropsychiatricdisorder, classified as an organic mental disorder and caused by the chronic meningoencephalitis that leads to cerebral atrophyin late-stage syphilis. Degenerative changes are associated primarily with the frontal and temporal lobar cortex. The disease affects approximately 7% of infected individuals. It is more common among men.
GPI was originally considered to be a type of madness due to a dissolute character, when first identified in the early 19th century. The condition's connection with syphilis was discovered in the late 1880s. Progressively, with the discovery of organic arsenicals such as Salvarsan and Neosalvarsan (1910s), the development of pyrotherapy (1920s), and the widespread availability and use of penicillin in the treatment of syphilis (1940s), the condition was rendered avoidable and curable. Prior to this, GPI was inevitably fatal, and it accounted for as much as 25% of the primary diagnoses for residents in public psychiatric hospitals.
Later: Delusions, dementia, tremors, hyperreflexia, seizures, cachexiaUsual onset10-30 years after initial infectionCausesMeningoencephalitis caused by syphilisRisk factorsUntreated syphilis infection
While retrospective studies have found earlier instances of what may have been the same disorder, the first clearly identified examples of paresis among the insane were described in Paris after the Napoleonic Wars. General paresis of the insane was first described as a distinct disease in 1822 by Antoine Laurent Jesse Bayle. General paresis most often struck people (men far more frequently than women) between 20 and 40 years of age. By 1877, for example, the superintendent of an asylum for men in New York reported that in his institution this disorder accounted for more than 12% of admissions and more than 2% of deaths.
Originally, the cause was believed to be an inherent weakness of character or constitution. While Friedrich von Esmarch and the psychiatrist Peter Willers Jessen had asserted as early as 1857 that syphilis caused general paresis (progressive Paralyse),[1] progress toward the general acceptance by the medical community of this idea was only accomplished later by the eminent 19th Century syphilographer Alfred Fournier (1832–1914). In 1913 all doubt about the syphilitic nature of paresis was finally eliminated when Hideyo Noguchi and J. W. Moore demonstrated the syphilitic spirochaetes in the brains of paretics.
In 1917 Julius Wagner-Jauregg discovered that pyrotherapy involving infecting paretic patients with malaria could halt the progression of general paresis. He won a Nobel Prize for this discovery in 1927. After World War II the use of penicillin to treat syphilis made general paresis a rarity: even patients manifesting early symptoms of actual general paresis were capable of full recovery with a course of penicillin. The disorder is now virtually unknown outside developing countries, and even there the epidemiology is substantially reduced.
Theo Van Gogh, brother of painter Vincent Van Gogh, died six months after Vincent in 1891 from "dementia parylitica" or what is now called syphilitic paresis.[2]
The Chicago gangster Al Capone died of syphilitic paresis, having contracted syphilis in a brothel prior to Prohibition and the Volstead Act and not having been treated for it in time to prevent the development of syphilitic paresis in himself.
https://en.wikipedia.org/wiki/General_paresis_of_the_insane
Neurosyphilis refers to infection of the central nervous system in a patient with syphilis. In the era of modern antibioticsthe majority of neurosyphilis cases have been reported in HIV-infected patients. Meningitis is the most common neurological presentation in early syphilis. Tertiary syphilis symptoms are exclusively neurosyphilis, though neurosyphilis may occur at any stage of infection.
To diagnose neurosyphilis, patients undergo a lumbar puncture to obtain cerebrospinal fluid (CSF) for analysis. The CSF is tested for antibodies for specific Treponema pallidum antigens. The preferred test is the VDRL test, which is sometimes supplemented by fluorescent treponemal antibody absorption test (FTA-ABS).[1][2][3]
Historically, the disease was studied under the Tuskegee study, a notable example of unethical human experimentation. The study was done on approximately 400 African-American men with untreated syphilis who were followed from 1932 to 1972 and compared to approximately 200 men without syphilis. The study began without informed consent of the subjects and was continued by the United States Public Health Service until 1972. The researchers failed to notify and withheld treatment for patients despite knowing penicillin was found as an effective cure for neurosyphilis. After four years of follow up, neurosyphilis was identified in 26.1% of patients vs. 2.5% of controls. After 20 years of followup, 14% showed signs of neurosyphilis and 40% had died from other causes.
Gummatous disease may also present with destructive inflammation and space-occupying lesions. It is caused by granulomatous destruction of visceral organs. They most often involve the frontal and parietal lobes of the brain.[citation needed]
https://en.wikipedia.org/wiki/Neurosyphilis
Shallot latent virus (SLV), a species of Carlavirus, was first identified in shallots in Netherlands.[1] The virus particle is elongated, 650 nm in length.
| Shallot latent virus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Kitrinoviricota |
| Class: | Alsuviricetes |
| Order: | Tymovirales |
| Family: | Betaflexiviridae |
| Genus: | Carlavirus |
| Species: | Shallot latent virus |
Since its first detection in shallots, SLV has been found infecting garlic, onion, and leek on five continents.[2][3][4][5][6]In Indonesia, the virus has been identified in shallot, which is widely grown and used as a food ingredient, and also in garlic.[7] In Turkey, where shallot is less commonly cultivated, SLV was identified in onion in Amasya province instead.[8] However, SLV was not detected in onion samples collected in Ankara province.[9] Molecular study also detected SLV in other Allium species such as Allium cyathophorum, Allium moly, Allium scorodoprasum, and Allium senescens subsp. montanum.[10]
The virus is widespread in shallot and garlic without causing any clear symptoms, hence its name 'latent'. However, in mixed infection with leek yellow stripe virus (LYSV, Potyvirus) induces severe chlorotic and white stripes on shallot leaves.[1] The aphids Myzus ascalonicus and Aphis fabae transmit SLV in a non-persistent manner, but Myzus persicae does not transmit the virus.[1] It is also mechanically transmitted.
https://en.wikipedia.org/wiki/Shallot_latent_virus
Myzus persicae, known as the green peach aphid, greenfly, or the peach-potato aphid,[2] is a small green aphid belonging to the order Hemiptera. It is the most significant aphid pest of peach trees, causing decreased growth, shrivelling of the leaves and the death of various tissues. It also acts as a vector for the transport of plant viruses such as cucumber mosaic virus (CMV), potato virus Y (PVY) and tobacco etch virus (TEV). Potato virus Y and potato leafroll virus can be passed to members of the nightshade/potato family (Solanaceae), and various mosaic viruses to many other food crops.[3]
Originally described by Swiss entomologist Johann Heinrich Sulzer in 1776, its specific name is derived from the Latin genitive persicae "of the peach".[4]
| Myzus persicae | |
|---|---|
 | |
| Scientific classification | |
| Kingdom: | Animalia |
| Phylum: | Arthropoda |
| Class: | Insecta |
| Order: | Hemiptera |
| Suborder: | Sternorrhyncha |
| Family: | Aphididae |
| Genus: | Myzus |
| Species: | M. persicae |
| Binomial name | |
| Myzus persicae | |
https://en.wikipedia.org/wiki/Myzus_persicae
Platysulcidae is a monotypic family of heterokonts that was recently discovered to be the earliest diverging lineage of the Heterokont phylogenetic tree.[1]
https://en.wikipedia.org/wiki/Platysulcidae
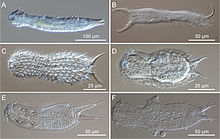

















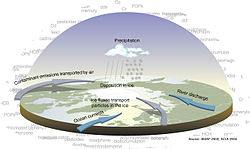















No comments:
Post a Comment