Above. Benny Goodman - Sing Sing Sing
-----------------------------------------------------------------------------------------------------------------------------
Annular elastolytic giant-cell granuloma (also known as "Giant cell elastophagocytosis,"[1]"Meischer's granuloma,"[2] "Miescher's granuloma of the face"[1]) is a cutaneous condition characterized histologically by a dermal infiltrate of macrophages.[1][2]:706
https://en.wikipedia.org/wiki/Annular_elastolytic_giant-cell_granuloma
Actinic granuloma is a cutaneous condition characterized histologically by a dermal infiltrate of macrophages.[1]:706
Actinic granuloma is an asymptomatic granulomatous reaction that affects sun-exposed skin, most commonly on the face, neck, and scalp. It is characterized by annular or polycyclic lesions that slowly expand centrifugally and have an erythematous elevated edge and a hypopigmented, atrophic center. Advise to reduce exposure to the sun and to use sunscreen. Treatment with topical halometasone cream, pimecrolimus cream.
https://en.wikipedia.org/wiki/Actinic_granuloma
Silica granulomas are a skin condition which may be caused by automobile and other types of accidents which produces tattooing of dirt (silicon dioxide) into the skin that then induces the granulomaformation.[1]:46
https://en.wikipedia.org/wiki/Silica_granuloma
Lichen planus (LP) is a chronic inflammatory and immune-mediated disease that affects the skin, nails, hair, and mucous membranes.[1] It is not an actual lichen, and is only named that because it looks like one.[2] It is characterized by polygonal, flat-topped, violaceous papules and plaques with overlying, reticulated, fine white scale (Wickham's striae), commonly affecting dorsal hands, flexural wrists and forearms, trunk, anterior lower legs and oral mucosa.[3] Although there is a broad clinical range of LP manifestations, the skin and oral cavity remain as the major sites of involvement.[4] The cause is unknown, but it is thought to be the result of an autoimmune process with an unknown initial trigger. There is no cure, but many different medications and procedures have been used in efforts to control the symptoms.
The term lichenoid reaction (lichenoid eruption or lichenoid lesion) refers to a lesion of similar or identical histopathologic and clinical appearance to lichen planus (i.e., an area which resembles lichen planus, both to the naked eye and under a microscope).[5][6] Sometimes dental materials or certain medications can cause a lichenoid reaction.[5] They can also occur in association with graft versus host disease.[5][7]:258
https://en.wikipedia.org/wiki/Lichen_planus#Classification
Tumid lupus erythematosus is a rare, but distinctive entity in which patients present with edematous erythematous plaques, usually on the trunk.[2]
Lupus erythematosus tumidus (LET) was reported by Henri Gougerot and Burnier R. in 1930. It is a photosensitive skin disorder, a different subtype of cutaneous lupus erythematosus (CLE) from discoid lupus erythematosus (DLE) or subacute CLE (SCLE).[3] LET is usually found on sun-exposed areas of the body. Skin lesions are edematous, urticarialike annular papules and plaques. Topical corticosteroids are not effective as treatment for LET, but many will respond to chloroquine. LET resolves with normal skin, no residual scarring, no hyperpigmentation or hypopigmentation. Cigarette smokers who have LET may not respond very well to chloroquine.[4][5]
It has been suggested that it is equivalent to Jessner lymphocytic infiltrate of the skin.[6]
https://en.wikipedia.org/wiki/Tumid_lupus_erythematosus
Verrucous lupus erythematosus presents with non-pruritic papulonodular lesions on the arms and hands, resembling keratoacanthoma or hypertropic lichen planus.[1]
https://en.wikipedia.org/wiki/Verrucous_lupus_erythematosus
Morphea, is a form of scleroderma that involves isolated patches of hardened skin on the face, hands, and feet, or anywhere else on the body, with no internal organ involvement.[1]:130
https://en.wikipedia.org/wiki/Morphea
Pyoderma gangrenosum is a rare, inflammatory skin disease where painful pustules or nodulesbecome ulcers that progressively grow.[3] Pyoderma gangrenosum is not infectious.[3]
Treatments may include corticosteroids, ciclosporin, infliximab, or canakinumab.[2]
The disease was identified in 1930. It affects approximately 1 person in 100,000 in the population. Though it can affect people of any age, it mostly affects people in their 40s and 50s.[1]
https://en.wikipedia.org/wiki/Pyoderma_gangrenosum
Poikiloderma vasculare atrophicans (PVA), is a cutaneous condition (skin disease) characterized by hypo- or hyperpigmentation (diminished or heightened skin pigmentation, respectively), telangiectasia and skin atrophy.[3][4][5] Other names for the condition include prereticulotic poikiloderma and atrophic parapsoriasis.[6] The condition was first described by pioneer American pediatrician Abraham Jacobi in 1906.[7] PVA causes areas of affected skin to appear speckled red and inflamed, yellowish and/or brown, gray or grayish-black, with scaling and a thinness that may be described as "cigarette paper".[3] On the surface of the skin, these areas may range in size from small patches, to plaques (larger, raised areas), to neoplasms(spreading, tumor-like growths on the skin).[3][6]
Mycosis fungoides, a type of skin lymphoma, may be a cause of PVA. The condition may also be caused by, associated with or accompany any of the following conditions or disorders: other skin lymphomas, dermatomyositis, lupus erythematosus, Rothmund–Thomson syndrome, Kindler syndrome, dyskeratosis congenita, and chronic radiodermatitis.[4] Rare causes include arsenicingestion, and the condition can also be idiopathic.[1][3][5]
PVA may be considered a rare variant of cutaneous T-cell lymphoma, a non-Hodgkin's form of lymphoma affecting the skin.[7] It may also be included among a number of similar conditions that are considered as precursors to mycosis fungoides. PVA is believed to be a syndrome closely associated with large-plaque parapsoriasis and its cohort retiform parapsoriasis; including PVA, all three conditions fit within an updated view of the once ambiguous classification scheme known as parapsoriasis.[5]
https://en.wikipedia.org/wiki/Poikiloderma_vasculare_atrophicans
Calcinosis cutis is a type of calcinosis wherein calcium deposits form in the skin. A variety of factors can result in this condition. The most common source is dystrophic calcification, which occurs in soft tissue as a response to injury. In addition, calcinosis is seen in Limited Cutaneous Systemic Sclerosis, also known as CREST syndrome (the "C" in CREST). In dogs, calcinosis cutis is found in young, large breed dogs and is thought to occur after a traumatic injury.
https://en.wikipedia.org/wiki/Calcinosis_cutis
Dystrophic calcification (DC) is the calcification occurring in degenerated or necrotic tissue, as in hyalinizedscars, degenerated foci in leiomyomas, and caseous nodules. This occurs as a reaction to tissue damage,[1]including as a consequence of medical device implantation. Dystrophic calcification can occur even if the amount of calcium in the blood is not elevated (a systemic mineral imbalance would elevate calcium levels in the blood and all tissues) and cause metastatic calcification. Basophilic calcium salt deposits aggregate, first in the mitochondria, then progressively throughout the cell. These calcifications are an indication of previous microscopic cell injury, occurring in areas of cell necrosis when activated phosphatases bind calcium ions to phospholipids in the membrane.
Calcification in dead tissue[edit]
- Caseous necrosis in T.B. is most common site of dystrophic calcification.
- Liquefactive necrosis in chronic abscesses may get calcified.
- Fat necrosis following acute pancreatitis or traumatic fat necrosis in breasts results in deposition of calcium soaps.
- Infarcts may undergo D.C.
- Thrombi, especially in veins, may produce phlebolithis.
- Haematomas in the vicinity of bones may undergo D.C.
- Dead parasites like schistosoma eggs may calcify.
- Congenital toxoplasmosis, CMV or rubella may be seen on X-ray as calcifications in the brain.
Calcification in degenerated tissue[edit]
- Dense scars may undergo hyaline degeneration and calcification.
- Atheroma in aorta and coronaries frequently undergo calcification.[2][3]
- Cysts can show calcification.
- Calcinosis cutis is condition in which there are irregular nodular deposits of calcium salts in skin and subcutaneous tissue.
- Senile degenerative changes may be accompanied by calcification.
- The inherited disorder pseudoxanthoma elasticum may lead to angioid streaks with calcification of Bruch's membrane, the elastic tissue below the retinal ring.
See also[edit]
Calciphylaxis, also known as calcific uremic arteriolopathy (CUA) or “Grey Scale”, is a rare painful syndrome of calcification of the small blood vessels located within the fatty tissue and deeper layers of the skin, blood clots, and the death of skin cells due to too little blood flow.[1] It is seen mostly in people with end-stage kidney disease but can occur in the earlier stages of chronic kidney disease and rarely in people with normally functioning kidneys.[1] It results in chronic non-healing wounds and is usually fatal. Calciphylaxis is a rare but serious disease, believed to affect 1-4% of all dialysis patients.[2]
Calciphylaxis is one type of extraskeletal calcification. Similar extraskeletal calcifications are observed in some people with high levels of calcium in the blood, including people with milk-alkali syndrome, sarcoidosis, primary hyperparathyroidism, and hypervitaminosis D. Certain medications such as warfarin can also result in calciphylaxis in rare cases. The presence of calciphylaxis generally predicts a poor prognosis with a typical life expectancy of less than one year.[1]
https://en.wikipedia.org/wiki/Calciphylaxis
Mönckeberg's arteriosclerosis, or Mönckeberg's sclerosis, is a form of arteriosclerosis or vessel hardening, where calcium deposits are found in the muscular middle layer of the walls of arteries (the tunica media).[1] It is an example of dystrophic calcification. This condition occurs as an age-related degenerative process. However, it can occur in pseudoxanthoma elasticum and idiopathic arterial calcification of infancy as a pathological condition, as well. Its clinical significance and cause are not well understood and its relationship to atherosclerosis and other forms of vascular calcification are the subject of disagreement.[2] Mönckeberg's arteriosclerosis is named after Johann Georg Mönckeberg,[3][4] who first described it in 1903.
https://en.wikipedia.org/wiki/Monckeberg%27s_arteriosclerosis
NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) is a protein complex that controls transcription of DNA, cytokineproduction and cell survival. NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, heavy metals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens.[2][3][4][6][7] NF-κB plays a key role in regulating the immune response to infection. Incorrect regulation of NF-κB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. NF-κB has also been implicated in processes of synaptic plasticity and memory.[8][9][10][11][12][13]
https://en.wikipedia.org/wiki/NF-κB
Dystrophic calcification (DC) is the calcification occurring in degenerated or necrotic tissue, as in hyalinizedscars, degenerated foci in leiomyomas, and caseous nodules. This occurs as a reaction to tissue damage,[1]including as a consequence of medical device implantation. Dystrophic calcification can occur even if the amount of calcium in the blood is not elevated (a systemic mineral imbalance would elevate calcium levels in the blood and all tissues) and cause metastatic calcification. Basophilic calcium salt deposits aggregate, first in the mitochondria, then progressively throughout the cell. These calcifications are an indication of previous microscopic cell injury, occurring in areas of cell necrosis when activated phosphatases bind calcium ions to phospholipids in the membrane.
https://en.wikipedia.org/wiki/Dystrophic_calcification
Generalized vaccinia is a cutaneous condition that occurs 6–9 days after vaccination, characterized by a generalized eruption of skin lesions, and caused by the vaccinia virus.[1]:391
https://en.wikipedia.org/wiki/Generalized_vaccinia
Progressive vaccinia is a rare cutaneous condition caused by the vaccinia virus, characterized by painless, but progressive, necrosis and ulceration.[4]
https://en.wikipedia.org/wiki/Progressive_vaccinia
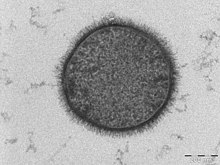
Vaccinia virus (VACV or VV) is a large, complex, enveloped virus belonging to the poxvirus family.[2] It has a linear, double-stranded DNA genome approximately 190 kbp in length, which encodes approximately 250 genes. The dimensions of the virion are roughly 360 × 270 × 250 nm, with a mass of approximately 5–10 fg.[3]The vaccinia virus is the source of the modern smallpox vaccine, which the World Health Organisation used to eradicate smallpox in a global vaccination campaign in 1958–1977. Although smallpox no longer exists in the wild, vaccinia virus is still studied widely by scientists as a tool for gene therapy and genetic engineering.
https://en.wikipedia.org/wiki/Vaccinia
Palmoplantar keratodermas are a heterogeneous group of disorders characterized by abnormal thickening of the stratum corneum of the palms and soles.
Autosomal recessive, dominant, X-linked, and acquired forms have all been described.[1]:505[2]:211[3]
| Palmoplantar keratoderma | |
|---|---|
 | |
| Patient with severe plantar keratosis. | |
| Specialty | Dermatology |
https://en.wikipedia.org/wiki/Palmoplantar_keratoderma
Molluscum contagiosum (MC), sometimes called water warts, is a viral infection of the skinthat results in small raised pink lesions with a dimple in the center.[1] They may become itchy or sore, and occur singularly or in groups.[1] Any area of the skin may be affected, with abdomen, legs, arms, neck, genital area, and face being the most common.[1] Onset of the lesions is around seven weeks after infection.[3] They usually go away within a year without scarring.[1]
https://en.wikipedia.org/wiki/Molluscum_contagiosum
Molluscum contagiosum virus (MCV) is a DNA poxvirus that causes the human skin infection molluscum contagiosum.[1] Molluscum contagiosum affects about 200,000 people a year, about 1% of all diagnosed skin diseases. Diagnosis is based on the size and shape of the skin lesions and can be confirmed with a biopsy, as the virus cannot be routinely cultured.[2] Molluscum contagiosum virus is the only species in the genus Molluscipoxvirus.[3] MCV is a member of the subfamily Chordopoxvirinae of family Poxviridae.[4] Other commonly known viruses that reside in the subfamily Chordopoxvirinae are variola virus (cause of smallpox) and monkeypox virus.[3]
The poxvirus family uniquely contains both non-enveloped particles (mature virions), and enveloped particles (extracellular virions).[5] The structure of the virions is consistent with that of others in the poxvirus family: they are composed of a nucleocapsid, core envelope, lateral body, and an extracellular envelope. Like other poxviruses, MCV is a DNA virus that replicates in the cytoplasm instead of the nucleus. Because of this, the virus must bring all necessary enzymes for replication with it or encode the enzymes in its genome.
- Wart (caused by the Human papillomavirus; also similar in appearance to molluscum)
https://en.wikipedia.org/wiki/Molluscum_contagiosum_virus
https://en.wikipedia.org/wiki/Category:Papillomavirus-associated_diseases
https://en.wikipedia.org/wiki/Basal-cell_carcinoma
https://en.wikipedia.org/wiki/Herpesviridae
Warts are typically small, rough, hard growths that are similar in color to the rest of the skin.[1][3]They typically do not result in other symptoms, except when on the bottom of the feet, where they may be painful.[3] While they usually occur on the hands and feet, they can also affect other locations.[1] One or many warts may appear.[3] They are not cancerous.[3]
Warts are caused by infection with a type of human papillomavirus (HPV).[1] Factors that increase the risk include use of public showers and pools, working with meat, eczema and a weak immune system.[1][3] The virus is believed to enter the body through skin that has been damaged slightly.[1] A number of types exist, including "common warts", plantar warts, "filiform warts", and genital warts.[3] Genital warts are often sexually transmitted.[5]
Warts are caused by the human papilloma virus (HPV)... dysplasia – "high-risk" HPV types are associated with cancers,...Verruca plana (flat warts) – HPV types 3, 10, and 28....Heck's disease (focal epithelial hyperplasia) – HPV types 13 and 32....Surviving ancient medical texts show that warts were a documented disease since at least the time of Hippocrates, who lived ca. 460 – c. 370 BC. In the book De Medecia by the Roman physician Aulus Cornelius Celsus, who lived c. 25 BC – c. 50 AD, different types of warts were described. Celsus described myrmecia, today recognized as plantar wart, and categorized acrochordon (a skin tag) as wart...Verrucae,[1] papillomas[2]
- Karamanou, Marianna; Agapitos, Emmanovil; Kousoulis, Antonis; Androutsos, George (17 August 2010). "From the humble wart to HPV: a fascinating story throughout centuries". Oncology Reviews. 4 (3): 133–135. doi:10.1007/s12156-010-0060-1. S2CID 72238300.
Fungal, Viral (with protein granulo viron or not), Parasite - crustacean, arthropod parsite, mite, plankton, mollusk, mosquito, protazo, protist, amoebae, trans-ge-lement, viral vector, gen-mod vector, gene transplant vector, DIW, amnestic drug, etc..
https://en.wikipedia.org/wiki/Wart
Eczema vaccinatum is a rare severe adverse reaction to smallpox vaccination.
It is characterized by serious local or disseminated, umbilicated, vesicular, crusting skin rashes in the face, neck, chest, abdomen, upper limbs and hands, caused by widespread infection of the skin in people with previous diagnosed skin conditions such as eczema or atopic dermatitis, even if the conditions are not active at the time. Other signs and symptoms include fever and facial and supraglottic edema. The condition may be fatal if severe and left untreated. Survivors are likely to have some scarring (pockmarks).
https://en.wikipedia.org/wiki/Eczema_vaccinatum
Minimally differentiated acute myeloblastic leukemia is a subtype of AML. It is classified as M0 by FAB. It represents 2–3% of all cases of AML.[1] Although minimally differentiated AML was recognized earlier, criteria for FAB M0 were developed in 1987.[2] The blasts in these cases cannot be recognized as myeloid based on morphology and cytochemistry, but immunophenotyping demonstrates myeloid antigens.[3]
https://en.wikipedia.org/wiki/Minimally_differentiated_acute_myeloblastic_leukemia
Pseudomonas aeruginosa is a common encapsulated, Gram-negative, strict aerobic (although can grow anaerobically in the presence of nitrate), rod-shaped bacterium that can cause disease in plants and animals, including humans.[1][2] A species of considerable medical importance, P. aeruginosa is a multidrug resistantpathogen recognized for its ubiquity, its intrinsically advanced antibiotic resistance mechanisms, and its association with serious illnesses – hospital-acquired infections such as ventilator-associated pneumonia and various sepsis syndromes.
https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa
Leukemia, also known as leukaemia, is a group of blood cancers that usually begin in the bone marrow and result in high numbers of abnormal blood cells.[9] These blood cells are not fully developed and are called blasts or leukemia cells.[2] Symptoms may include bleeding and bruising, bone pain, fatigue, fever, and an increased risk of infections.[2] These symptoms occur due to a lack of normal blood cells.[2] Diagnosis is typically made by blood tests or bone marrow biopsy.[2]
https://en.wikipedia.org/wiki/Leukemia
Methotrexate (MTX), formerly known as amethopterin, is a chemotherapy agent and immune-system suppressant.[4] It is used to treat cancer, autoimmune diseases, and ectopic pregnancyand for medical abortions.[4] Types of cancers it is used for include breast cancer, leukemia, lung cancer, lymphoma, gestational trophoblastic disease, and osteosarcoma.[4] Types of autoimmune diseases it is used for include psoriasis, rheumatoid arthritis, and Crohn's disease.[4] It can be given by mouth or by injection.[4]
https://en.wikipedia.org/wiki/Methotrexate
Myxococcus is a genus in the family Myxococcaceae. Myxococci are Gram-negative, spore-forming, chemoorganotrophic, obligate aerobes. They are elongated rods with rounded or tapered ends, and they are nonflagellated. The cells move by gliding and can predate other bacteria. The genus has been isolated from soil.
https://en.wikipedia.org/wiki/Myxococcus
The human T-lymphotropic virus, human T-cell lymphotropic virus, or human T-cell leukemia-lymphoma virus (HTLV) family of viruses are a group of human retroviruses that are known to cause a type of cancer called adult T-cell leukemia/lymphoma and a demyelinating disease called HTLV-1 associated myelopathy/tropical spastic paraparesis (HAM/TSP). The HTLVs belong to a larger group of primate T-lymphotropic viruses (PTLVs). Members of this family that infect humans are called HTLVs, and the ones that infect Old World monkeys are called Simian T-lymphotropic viruses (STLVs). To date, four types of HTLVs (HTLV-1, HTLV-2, HTLV-3, and HTLV-4) and four types of STLVs (STLV-1, STLV-2, STLV-3, and STLV-5) have been identified. HTLV types HTLV-1 and HTLV-2 viruses are the first retroviruses discovered. Both belong to the oncovirus subfamily of retroviruses and can transform human lymphocytes so that they are self-sustaining in vitro.[1] The HTLVs are believed to originate from interspecies transmission of STLVs. The HTLV-1 genome is diploid, composed of two copies of a single-stranded RNA virus whose genome is copied into a double-stranded DNA form that integrates into the host cell genome, at which point the virus is referred to as a provirus. A closely related virus is bovine leukemia virus BLV. The original name for HIV, the virus that causes AIDS, was HTLV-3.
https://en.wikipedia.org/wiki/Human_T-lymphotropic_virus
Visna-maedi virus (also known as Visna virus, Maedi-visna virus and Ovine lentivirus[1]) from the genusLentivirus and subfamily Orthoretrovirinae, is a retrovirus that causes encephalitis and chronic pneumonitis in sheep.[2][3][4] It is known as visna when found in the brain, and maedi when infecting the lungs. Lifelong, persistent infections in sheep occur in the lungs, lymph nodes, spleen, joints, central nervous system, and mammary glands;[2][5] The condition is sometimes known as ovine progressive pneumonia (OPP), particularly in the United States,[1] or Montana sheep disease.[6] White blood cells of the monocyte/macrophage lineage are the main target of the virus.[7]
https://en.wikipedia.org/wiki/Visna-maedi_virus
Bacillus thuringiensis (or Bt) is a Gram-positive, soil-dwelling bacterium, the most commonly used biological pesticide worldwide. B. thuringiensis also occurs naturally in the gut of caterpillars of various types of mothsand butterflies, as well on leaf surfaces, aquatic environments, animal feces, insect-rich environments, and flour mills and grain-storage facilities.[1][2] It has also been observed to parasitize other moths such as Cadra calidella—in laboratory experiments working with C. calidella, many of the moths were diseased due to this parasite.[3]
https://en.wikipedia.org/wiki/Bacillus_thuringiensis
Lentivirus is a genus of retroviruses that cause chronic and deadly diseases characterized by long incubation periods, in humans and other mammalian species.[1] The genus includes the human immunodeficiency virus(HIV), which causes AIDS. Lentiviruses are distributed worldwide, and are known to be hosted in apes, cows, goats, horses, cats, and sheep as well as several other mammals.[1]
Lentiviruses can integrate a significant amount of viral complementary DNA into the DNA of the host cell and can efficiently infect nondividing cells, so they are one of the most efficient methods of gene delivery.[2] They can become endogenous, integrating their genome into the host germline genome, so that the virus is henceforth inherited by the host's descendants.
https://en.wikipedia.org/wiki/Lentivirus
Tumor lysis syndrome is a group of metabolic abnormalities that can occur as a complication during the treatment of cancer,[1] where large amounts of tumor cells are killed off (lysed) at the same time by the treatment, releasing their contents into the bloodstream. This occurs most commonly after the treatment of lymphomas and leukemias. In oncology and hematology, this is a potentially fatal complication, and patients at increased risk for TLS should be closely monitored before, during, and after their course of chemotherapy.
Tumor lysis syndrome is characterized by high blood potassium (hyperkalemia), high blood phosphate (hyperphosphatemia), low blood calcium (hypocalcemia), high blood uric acid (hyperuricemia), and higher than normal levels of blood urea nitrogen (BUN) and other nitrogen-containing compounds (azotemia). These changes in blood electrolytes and metabolites are a result of the release of cellular contents of dying cells into the bloodstream from breakdown of cells. In this respect, TLS is analogous to rhabdomyolysis, with comparable mechanism and blood chemistry effects but with different cause. In TLS, the breakdown occurs after cytotoxic therapy or from cancers with high cell turnover and tumor proliferation rates. The metabolic abnormalities seen in tumor lysis syndrome can ultimately result in nausea and vomiting, but more seriously acute uric acid nephropathy, acute kidney failure, seizures, cardiac arrhythmias, and death.[2][3]
https://en.wikipedia.org/wiki/Tumor_lysis_syndrome
Tumor necrosis factor (TNF, cachexin, or cachectin; often called tumor necrosis factor alpha or TNF-α) is a cytokine – a small protein used by the immune system for cell signaling. If macrophages (certain white blood cells) detect an infection, they release TNF to alert other immune system cells as part of an inflammatory response. TNF is a member of the TNF superfamily, which consists of various transmembrane proteins with a homologous TNF domain.
https://en.wikipedia.org/wiki/Tumor_necrosis_factor
Rhabdomyolysis is a condition in which damaged skeletal muscle breaks down rapidly.[3]Symptoms may include muscle pains, weakness, vomiting, and confusion.[3][4] There may be tea-colored urine or an irregular heartbeat.[3] Some of the muscle breakdown products, such as the protein myoglobin, are harmful to the kidneys and may lead to kidney failure.[3]
https://en.wikipedia.org/wiki/Rhabdomyolysis
Caprine arthritis encephalitis virus (CAEV) is a retrovirus which infects goats and cross-reacts immunologically with HIV,[1] due to being from the same family of viruses.[2] CAEV cannot be transmitted to humans, including through the consumption of milk from an infected goat.[3] There is no evidence that CAEV can cure HIV in humans.[2][4]
CAEV is commonly transferred within the goat species by ingestion of colostrum or milk from an infected goat, and to a less extent, cross species CAEV transfer by sheep is possible.[3][5]
https://en.wikipedia.org/wiki/Caprine_arthritis_encephalitis_virus
Jaagsiekte sheep retrovirus (JSRV) is a betaretrovirus which is the causative agent of a contagious lung cancer in sheep, called ovine pulmonary adenocarcinoma.
https://en.wikipedia.org/wiki/Jaagsiekte_sheep_retrovirus
Simian immunodeficiency virus (SIV) is a species of retrovirus that cause persistent infections in at least 45 species of African non-human primates.[1][2] Based on analysis of strains found in four species of monkeysfrom Bioko Island, which was isolated from the mainland by rising sea levels about 11,000 years ago, it has been concluded that SIV has been present in monkeys and apes for at least 32,000 years, and probably much longer.[3][4]
https://en.wikipedia.org/wiki/Simian_immunodeficiency_virus
Treponema pallidum is a spirochaete bacterium with various subspecies that cause the diseases syphilis, bejel, and yaws. It is transmitted only amongst humans.[1] It is a helically coiled microorganism usually 6–15 μm long and 0.1–0.2 μm wide.[2] T. pallidum's lack of either tricarboxylic acid cycle or oxidative phosphorylation results in minimal metabolic activity.[3] The treponemes have a cytoplasmic and an outer membrane. Using light microscopy, treponemes are visible only by using dark field illumination. Treponema pallidum consists of three subspecies, T. p. pallidum, T. p. endemicum, and T. p. pertenue, each of which has a distinct associated disease.[4]
https://en.wikipedia.org/wiki/Treponema_pallidum
Syphilis is a sexually transmitted infection caused by the bacterium Treponema pallidumsubspecies pallidum.[3] The signs and symptoms of syphilis vary depending in which of the four stages it presents (primary, secondary, latent, and tertiary).[1] The primary stage classically presents with a single chancre (a firm, painless, non-itchy skin ulceration usually between 1 cm and 2 cm in diameter) though there may be multiple sores.[1] In secondary syphilis, a diffuse rashoccurs, which frequently involves the palms of the hands and soles of the feet.[1] There may also be sores in the mouth or vagina.[1] In latent syphilis, which can last for years, there are few or no symptoms.[1] In tertiary syphilis, there are gummas (soft, non-cancerous growths), neurological problems, or heart symptoms.[2] Syphilis has been known as "the great imitator" as it may cause symptoms similar to many other diseases.[1][2]
https://en.wikipedia.org/wiki/Syphilis
Endogenous retroviruses (ERVs) are endogenous viral elements in the genome that closely resemble and can be derived from retroviruses. They are abundant in the genomes of jawed vertebrates, and they comprise up to 5–8% of the human genome (lower estimates of ~1%).[1][2]
https://en.wikipedia.org/wiki/Endogenous_retrovirus#Human_endogenous_retroviruses
A provirus is a virus genome that is integrated into the DNA of a host cell. In the case of bacterial viruses (bacteriophages), proviruses are often referred to as prophages. However, it is important to note that proviruses are distinctly different from prophages and these terms should not be used interchangeably. Unlike prophages, proviruses do not excise themselves from the host genome when the host cell is stressed.[1][page needed]
This state can be a stage of virus replication, or a state that persists over longer periods of time as either inactive viral infections or an endogenous viral element. In inactive viral infections the virus will not replicate itself except through replication of its host cell. This state can last over many host cell generations.
Endogenous retroviruses are always in the state of a provirus. When a (nonendogenous) retrovirus invades a cell, the RNA of the retrovirus is reverse-transcribed into DNA by reverse transcriptase, then inserted into the host genome by an integrase.
A provirus not only refers to a retrovirus but is also used to describe other viruses that can integrate into the host chromosomes, another example being adeno-associated virus. Not only eukaryotic viruses integrate into the genomes of their hosts; many bacterial and archaeal viruses also employ this strategy of propagation. All families of bacterial viruses with circular (single-stranded or double-stranded) DNA genomes or replicating their genomes through a circular intermediate (e.g., tailed dsDNA viruses) have temperate members.[5]
https://en.wikipedia.org/wiki/Provirus
A prophage is a bacteriophage (often shortened to "phage") genome inserted and integrated into the circular bacterial DNA chromosome or exists as an extrachromosomal plasmid. This is a latent form of a phage, in which the viral genes are present in the bacterium without causing disruption of the bacterial cell. Pro means "before", so, prophage means the stage of a virus in the form of genome inserted into host DNA before being activated inside the host.
https://en.wikipedia.org/wiki/Prophage
Bacillus thuringiensis (or Bt) is a Gram-positive, soil-dwelling bacterium, the most commonly used biological pesticide worldwide. B. thuringiensis also occurs naturally in the gut of caterpillars of various types of mothsand butterflies, as well on leaf surfaces, aquatic environments, animal feces, insect-rich environments, and flour mills and grain-storage facilities.[1][2] It has also been observed to parasitize other moths such as Cadra calidella—in laboratory experiments working with C. calidella, many of the moths were diseased due to this parasite.[3]
During sporulation, many Bt strains produce crystal proteins (proteinaceous inclusions), called delta endotoxins, that have insecticidal action. This has led to their use as insecticides, and more recently to genetically modified crops using Bt genes, such as Bt corn.[4] Many crystal-producing Bt strains, though, do not have insecticidal properties.[5] The subspecies israelensis is commonly used for control of mosquitoes[6]and of fungus gnats.[7]
As a toxic mechanism, cry proteins bind to specific receptors on the membranes of mid-gut (epithelial) cells of the targeted pests, resulting in their rupture. Other organisms (including humans, other animals and non-targeted insects) that lack the appropriate receptors in their gut cannot be affected by the cry protein, and therefore are not affected by Bt.[8][9]
B. thuringiensis is placed in the Bacillus cereus group which is variously defined as: seven closely related species: B. cereus sensu stricto (B. cereus), B. anthracis, B. thuringiensis, B. mycoides, B. pseudomycoides, and B. cytotoxicus;[17] or as six species in a Bacillus cereus sensu lato: B. weihenstephanensis, B. mycoides, B. pseudomycoides, B. cereus, B. thuringiensis, and B. anthracis. Within this grouping B.t. is more closely related to B.ce. It is more distantly related to B.w., B.m., B.p., and B.cy.[18]
Mechanism of insecticidal action[edit]
Upon sporulation, B. thuringiensis forms crystals of two types of proteinaceous insecticidal delta endotoxins (δ-endotoxins) called crystal proteins or Cry proteins, which are encoded by cry genes, and Cyt proteins.[23]
Cry toxins have specific activities against insect species of the orders Lepidoptera (moths and butterflies), Diptera (flies and mosquitoes), Coleoptera(beetles) and Hymenoptera (wasps, bees, ants and sawflies), as well as against nematodes.[27][28] Thus, B. thuringiensis serves as an important reservoir of Cry toxins for production of biological insecticides and insect-resistant genetically modified crops. When insects ingest toxin crystals, their alkaline digestive tracts denature the insoluble crystals, making them soluble and thus amenable to being cut with proteases found in the insect gut, which liberate the toxin from the crystal.[24] The Cry toxin is then inserted into the insect gut cell membrane, paralyzing the digestive tract and forming a pore.[29] The insect stops eating and starves to death; live Bt bacteria may also colonize the insect, which can contribute to death.[24][29][30] Death occurs within a few hours or weeks.[31] The midgut bacteria of susceptible larvae may be required for B. thuringiensis insecticidal activity.[32]
A B. thuringiensis small RNA called BtsR1 can silence the Cry5Ba toxin expression when outside the host by binding to the RBS site of the Cry5Ba toxin transcript to avoid nematode behavioral defenses. The silencing results in an increased of the bacteria ingestion by C. elegans.The expression of BtsR1 is then reduced after ingestion, resulting in Cry5Ba toxin production and host death.[33]
In 1996 another class of insecticidal proteins in Bt was discovered: the vegetative insecticidal proteins (Vip; InterPro: IPR022180).[34][35] Vip proteins do not share sequence homology with Cry proteins, in general do not compete for the same receptors, and some kill different insects than do Cry proteins.[34]
In 2000, a novel subgroup of Cry protein, designated parasporin, was discovered from non-insecticidal B. thuringiensis isolates.[36] The proteins of parasporin group are defined as B. thuringiensis and related bacterial parasporal proteins that are not hemolytic, but capable of preferentially killing cancer cells.[37] As of January 2013, parasporins comprise six subfamilies: PS1 to PS6.[38]
Use of spores and proteins in pest control[edit]
Spores and crystalline insecticidal proteins produced by B. thuringiensis have been used to control insect pests since the 1920s and are often applied as liquid sprays.[39] They are now used as specific insecticides under trade names such as DiPel and Thuricide. Because of their specificity, these pesticides are regarded as environmentally friendly, with little or no effect on humans, wildlife, pollinators, and most other beneficial insects, and are used in organic farming;[28] however, the manuals for these products do contain many environmental and human health warnings,[40][41] and a 2012 European regulatory peer review of five approved strains found, while data exist to support some claims of low toxicity to humans and the environment, the data are insufficient to justify many of these claims.[42]
New strains of Bt are developed and introduced over time[43] as insects develop resistance to Bt,[44] or the desire occurs to force mutations to modify organism characteristics[45][clarification needed], or to use homologous recombinant genetic engineering to improve crystal size and increase pesticidal activity,[46] or broaden the host range of Bt and obtain more effective formulations.[47] Each new strain is given a unique number and registered with the U.S. EPA[48] and allowances may be given for genetic modification depending on "its parental strains, the proposed pesticide use pattern, and the manner and extent to which the organism has been genetically modified".[49] Formulations of Bt that are approved for organic farming in the US are listed at the website of the Organic Materials Review Institute (OMRI)[50] and several university extension websites offer advice on how to use Bt spore or protein preparations in organic farming.[51][52]
Use of Bt genes in genetic engineering of plants for pest control[edit]
The Belgian company Plant Genetic Systems (now part of Bayer CropScience) was the first company (in 1985) to develop genetically modified crops(tobacco) with insect tolerance by expressing cry genes from B. thuringiensis; the resulting crops contain delta endotoxin.[53][54] The Bt tobacco was never commercialized; tobacco plants are used to test genetic modifications since they are easy to manipulate genetically and are not part of the food supply.[55][56]
Beta-exotoxins[edit]
Some isolates of B. thuringiensis produce a class of insecticidal small molecules called beta-exotoxin, the common name for which is thuringiensin.[118] A consensus document produced by the OECD says: "Beta-exotoxins are known to be toxic to humans and almost all other forms of life and its presence is prohibited in B. thuringiensis microbial products".[119] Thuringiensins are nucleoside analogues. They inhibit RNA polymeraseactivity, a process common to all forms of life, in rats and bacteria alike.[120]
Other hosts[edit]
Opportunistic pathogen of animals other than insects, causing necrosis, pulmonary infection, and/or food poisoning. How common this is is unknown because these are always taken to be B. cereus infections and are rarely tested for the Cry and Cyt proteins that are the only factor distinguishing .B thuringiensis from B. cereus.[18]
See also[edit]
https://en.wikipedia.org/wiki/Bacillus_thuringiensis
- Unintended consequences: Studies have found broad spectrum biopesticides have lethal and nonlethal risks for non-target native pollinators such as Melipona quadrifasciata in Brazil.[17]
Pages in category "Bacteria described in 1915"
The following 3 pages are in this category, out of 3 total. This list may not reflect recent changes (learn more).
B
D
Bacillus coagulans (Weizmannia coagulans) is a lactic acid–forming bacterial species first isolated and described in 1915 by B.W. Hammer at the Iowa Agricultural Experiment Station as a cause of an outbreak of coagulation in evaporated milk packed by an Iowa condensary.[1] Separately isolated in 1935 and described as Lactobacillus sporogenes in the fifth edition of Bergey's Manual of Systematic Bacteriology, it exhibits characteristics typical of both genera Lactobacillus and Bacillus; its taxonomic position between the families Lactobacillaceae and Bacillaceae was often debated. However, in the seventh edition of Bergey's, it was finally transferred to the genus Bacillus. DNA-based technology was used in distinguishing between the two genera of bacteria, which are morphologically similar and possess similar physiological and biochemicalcharacteristics.[2][3]
Bacillus coagulans is a Gram-positive, catalase-positive, spore-forming, motile, facultative anaerobe rod that measures approximately 0.9 μm by 3.0 μm to 5.0 μm. It may appear Gram negative when entering the stationary phase of growth. The optimum temperature for growth is 50 °C (122 °F); range of temperatures tolerated are 30–55 °C (86–131 °F). IMViC tests VP and MR (methyl red) are positive.
This species has been recently transferred into the genus Weizmannia.[4]
https://en.wikipedia.org/wiki/Bacillus_coagulans
Dermatophilus congolensis is a Gram-positive bacterium and the cause of a disease called dermatophilosis (sometimes called mud fever) in animals and humans, a dermatologic condition that manifests as the formation of crusty scabs containing the microorganism. It has been erroneously called mycotic dermatitis.[1] Rainscald is another condition often seen in animals, which is also caused by D. congolensis.
https://en.wikipedia.org/wiki/Dermatophilus_congolensis
Cytopathology (from Greek κύτος, kytos, "a hollow";[1] πάθος, pathos, "fate, harm"; and -λογία, -logia) is a branch of pathology that studies and diagnoses diseases on the cellular level. The discipline was founded by George Nicolas Papanicolaou in 1928. Cytopathology is generally used on samples of free cells or tissue fragments, in contrast to histopathology, which studies whole tissues. Cytopathology is frequently, less precisely, called "cytology", which means "the study of cells".[2]
Cytopathology is commonly used to investigate diseases involving a wide range of body sites, often to aid in the diagnosis of cancer but also in the diagnosis of some infectious diseases and other inflammatory conditions.[3] For example, a common application of cytopathology is the Pap smear, a screening tool used to detect precancerous cervical lesions that may lead to cervical cancer.
Cytopathologic tests are sometimes called smear tests because the samples may be smeared across a glass microscope slide[4] for subsequent staining and microscopic examination. However, cytology samples may be prepared in other ways, including cytocentrifugation. Different types of smear tests may also be used for cancer diagnosis. In this sense, it is termed a cytologic smear.[5]
https://en.wikipedia.org/wiki/Cytopathology
Various - cytoskeleton arthropod follicle tubule channel tube tuber protuberans macrophage filopodia filopod amoeba.
Human - ovulation ovary fertility fertilization etc.; Gene, DNA, genetic signature, gene code, genetics, etc.; embryology, biology, chemistry, physics, mathematics, language, etc..
Human papillomavirus infection (HPV infection) is an infection caused by human papillomavirus (HPV), a DNA virus from the Papillomaviridae family.[5] Many HPV infections cause no symptoms and 90% resolve spontaneously within two years.[1] However, in some cases, an HPV infection persists and results in either warts or precancerous lesions.[2] These lesions, depending on the site affected, increase the risk of cancer of the cervix, vulva, vagina, penis, anus, mouth, tonsils, or throat.[1][2][3] Nearly all cervical cancer is due to HPV; two strains, HPV16 and HPV18, account for 70% of cases.[1][7] HPV16 is responsible for almost 90% of HPV-positive oropharyngeal cancers.[3] Between 60% and 90% of the other cancers listed above are also linked to HPV.[7] HPV6 and HPV11 are common causes of genital warts and laryngeal papillomatosis.[1]
https://en.wikipedia.org/wiki/Human_papillomavirus_infection
Human endogenous retroviruses (HERV) comprise a significant part of the human genome, with approximately 98,000 ERV elements and fragments making up 5–8%.[1] According to a study published in 2005, no HERVs capable of replication had been identified; all appeared to be defective, containing major deletions or nonsense mutations. This is because most HERVs are merely traces of original viruses, having first integrated millions of years ago. An analysis of HERV integrations is ongoing as part of the 100,000 Genomes Project.[57]
Human endogenous retroviruses were discovered by accident using a couple of different experiments. Human genomic libraries were screened under low-stringency conditions using probes from animal retroviruses, allowing the isolation and characterization of multiple, though defective, proviruses, that represented various families. Another experiment depended on oligonucleotides with homology to viral primer binding sites.[1]
HERVs are classified based on their homologies to animal retroviruses. Families belong to Class I are similar in sequence to mammalian Gammaretroviruses (type C) and Epsilonretroviruses (Type E). Families belonging to Class II show homology to mammalian Betaretroviruses (Type B) and Deltaretroviruses(Type D). Families belong to Class III are similar to foamy viruses. For all classes, if homologies appear well conserved in the gag, pol, and env gene, they are grouped into a superfamily. There are more Class I families known to exist.[1][11] The families themselves are named in a less uniform manner, with a mixture of naming based on an exogenous retrovirus, the priming tRNA (HERV-W, K), or some neighboring gene (HERV-ADP), clone number (HERV-S71), or some amino acid motif (HERV-FRD). A proposed nomenclature aims to clean up the sometimes paraphyletic standards.[58]
There are two proposals for how HERVs became fixed in the human genome. The first assumes that sometime during human evolution, exogenous progenitors of HERV inserted themselves into germ line cells and then replicated along with the host's genes using and exploiting the host's cellular mechanisms. Because of their distinct genomic structure, HERVs were subjected to many rounds of amplification and transposition, which lead to a widespread distribution of retroviral DNA. The second hypothesis claims the continuous evolution of retro-elements from more simple structured ancestors.[1]
Nevertheless, one family of viruses has been active since the divergence of humans and chimpanzees. This family, termed HERV-K (HML2), makes up less than 1% of HERV elements but is one of the most studied. There are indications it has even been active in the past few hundred thousand years, e.g., some human individuals carry more copies of HML2 than others.[59] Traditionally, age estimates of HERVs are performed by comparing the 5' and 3' LTR of a HERV; however, this method is only relevant for full-length HERVs. A recent method, called cross-sectional dating,[60] uses variations within a single LTR to estimate the ages of HERV insertions. This method is more precise in estimating HERV ages and can be used for any HERV insertions. Cross-sectional dating has been used to suggest that two members of HERV-K(HML2), HERV-K106 and HERV-K116, were active in the last 800,000 years and that HERV-K106 may have infected modern humans 150,000 years ago.[61] However, the absence of known infectious members of the HERV-K(HML2) family, and the lack of elements with a full coding potential within the published human genome sequence, suggests to some that the family is less likely to be active at present. In 2006 and 2007, researchers working independently in France and the US recreated functional versions of HERV-K(HML2).[62][63]
https://en.wikipedia.org/wiki/Endogenous_retrovirus#Human_endogenous_retroviruses
Human endogenous retrovirus K (HERV-K) or Human teratocarcinoma-derived virus (HDTV) is a family of human endogenous retroviruses associated with malignant tumors of the testes.[1][2][3][4] Phylogenetically, the HERV-K group belongs to the ERV2 or Class II or Betaretrovirus-like supergroup.[5] Over the past several years, it has been found that this group of ERVs play an important role in embryogenesis, but their expression is silenced in most cell types in healthy adults.[6] The HERV-K family, and particularly its subgroup HML-2, is the youngest and most transcriptionally active group and hence, it is the best studied among other ERVs. Reactivation of it or anomalous expression of HML-2 in adult tissues has been associated with various types of cancer [7][8][9] and with neurodegenerative diseases such as amytrophic lateral sclerosis (ALS).[10][5]endogenous retrovirus K (HERV-K) is related to mammary tumor virus in mice. It exists in the human and cercopithecoid genomes. Human genome contains hundreds of copies of HERV-K and many of them possess complete open reading frames (ORFs) that are transcribed and translated, especially in early embryogenesis and in malignancies.[5][11] HERV-K is also found in apes and Old World monkeys. It is uncertain how long ago in primate evolution the full-length HERV-K proviruses which are in the human genome today were created.[12]
The human endogenous retrovirus K (HERV-K) was inherited million years ago by the genome of the human ancestors.[12] In 1999 Barbulescu, et al. showed that, of ten HERV-K proviruses cloned, eight were unique to humans, while one was shared with chimpanzees and bonobos, and one with chimpanzees, bonobos and gorillas.[13] Originally, HERV-K was observed by low-stringency hybridization with probes for the mammary tumor virus of the mouse and A particle intracutaneous mouse.[12]
In 2015 Grow et al. demonstrated that HERV-K is transcribed during embryogenesis from the eight cell stage up to the stem cell derivation.[14]Furthermore, overexpression of the HERV-K accessory protein Rec (regulator of expression encoded by corf; Pfam PF15695) increases IFITM1levels on the cell surface and inhibits viral infection.[14][15] HERV-K is called, phylogenetically, a supergroup of viruses. It is the only group that reported to contain human-specific members of endogenous retroviruses (ERVs).[16]
HERV-K is receptive to microenvironmental modifications and melanoma cells are closely correlated with epigenetic and microenvironmental anomalies. Also the association of HERV-K activation with carcinogenesis is especially interesting.[17]
https://en.wikipedia.org/wiki/Human_endogenous_retrovirus_K
Deltaretrovirus is a genus of the Retroviridae family. It consists of exogenous horizontally transmitted viruses found in several groups of mammals.
Examples are the Bovine leukemia virus and the human T-lymphotropic viruses.
https://en.wikipedia.org/wiki/Deltaretrovirus
Spumaretrovirinae, commonly called spumaviruses (spuma, Latin for "foam") or foamyviruses, is a subfamily of the Retroviridae family.[2] Spumaviruses are exogenous viruses that have specific morphology with prominent surface spikes. The virions contain significant amounts of double-stranded full-length DNA, and assembly is rather unusual in these viruses. Spumaviruses are unlike most enveloped viruses in that the envelope membrane is acquired by budding through the endoplasmic reticulum instead of the cytoplasmic membrane. Some spumaviruses, including the equine foamy virus (EFV), bud from the cytoplasmic membrane.
Some examples of these viruses are simian foamy virus and the human foamy virus.
While spumaviruses will form characteristic large vacuoles in their host cells while in vitro, there is no disease association in vivo.[3]
https://en.wikipedia.org/wiki/Spumaretrovirinae
Soon after BLV was discovered in the 1970s, ten studies were done looking for antibodies to BLV in humans. However, no antibodies were found and so researchers concluded that BLV was not a risk to human health.[8] However, more sensitive techniques for detecting antibodies were developed, and in 2003 a test of more than 200 people using these new tests found that more than a third carried antibodies reactive to BLV, and the question began to be researched again.[9]
Rabbits get a fatal AIDS-like disease similar to Pasteurella, different from the benign human snuffles.
https://en.wikipedia.org/wiki/Bovine_leukemia_virus
Rhinitis, also known as coryza,[3] is irritation and inflammation of the mucous membrane inside the nose. Common symptoms are a stuffy nose, runny nose, sneezing, and post-nasal drip.[4]
The inflammation is caused by viruses, bacteria, irritants or allergens. The most common kind of rhinitis is allergic rhinitis, [5] which is usually triggered by airborne allergens such as pollen and dander.[6] Allergic rhinitis may cause additional symptoms, such as sneezing and nasal itching, coughing, headache,[7] fatigue, malaise, and cognitive impairment.[8][9] The allergens may also affect the eyes, causing watery, reddened, or itchy eyes and puffiness around the eyes.[7] The inflammation results in the generation of large amounts of mucus, commonly producing a runny nose, as well as a stuffy nose and post-nasal drip. In the case of allergic rhinitis, the inflammation is caused by the degranulation of mast cells in the nose. When mast cells degranulate, they release histamine and other chemicals,[10] starting an inflammatory process that can cause symptoms outside the nose, such as fatigue and malaise.[11] In the case of infectious rhinitis, it may occasionally lead to pneumonia, either viral or bacterial. Sneezing also occurs in infectious rhinitis to expel bacteria and viruses from the respiratory tract.
Rhinitis is very common. Allergic rhinitis is more common in some countries than others; in the United States, about 10–30% of adults are affected annually.[12] Mixed rhinitis (MR) refers to patients with nonallergic rhinitis and allergic rhinitis. MR is a specific rhinitis subtype. It may represent between 50 and 70% of all AR patients. However, true prevalence of MR has not been confirmed yet.[13]
https://en.wikipedia.org/wiki/Rhinitis
Leucosis is a leukemia-like malignant viral disease that is found in animals, particularly poultry and cattle.
Types of leucosis[edit]
- Bovine leucosis
- Enzootic bovine leucosis, caused by bovine leukemia virus.[1]
- Sporadic bovine leucosis
- Calf lymphosarcoma
- Leucosis in pig
- Leucosis in horses
- Leucosis in sheep [2]
- Feline leucosis
- Avian leucosis and related diseases
- Avian sarcoma leukosis virus
- Lymphoid leucosis
- Erythroblastosis
- Osteopetrosis
- Myeloblastose
- Myelocytomatosis[3]
FeLV was first described in cats in 1964.[25] The disease was originally associated with leukemia; however, it was later realized that the initial signs are generally anemia and immunosuppression.[25] The first diagnostic test became available in 1973, which led to a "test and elimination" regime, dramatically reducing the number of infected cats in the general population.[26] The first vaccine became available in 1986.[26]
Progression
[edit]
The disease has a wide range of effects. The cat can fight off the infection and become totally immune, can become a healthy carrier that never gets sick itself but can infect other cats, or a mid-level case in which the cat has a compromised immune system.[citation needed] Nevertheless, the development of lymphomas is considered the final stage of the disease. Although it is thought that virus protein has to be present to induce lymphomas in cats, newer evidence shows that a high percentage of FeLV-Antigen negative lymphomas contain FeLV-DNA, indicating a "hit-and-run" mechanism of virus-induced tumor development.[5]
Once the virus has entered the cat, there are six stages to a FeLV infection[citation needed]:
- Stage One: The virus enters the cat, usually through the pharynx where it infects the epithelial cells and infects the tonsilar B-lymphocytes and macrophages. These white blood cells then filter down to the lymph nodes and begin to replicate.
- Stage Two: The virus enters the blood stream and begins to distribute throughout the body.
- Stage Three: The lymphoid system (which produces antibodies to attack infected and cancerous cells) becomes infected, with further distribution throughout the body.
- Stage Four: The main point in the infection- where the virus can take over the body's immune system and cause viremia. During this stage the hemolymphatic system and intestines become infected.
If the cat's immune system does not fight off the virus, then it progresses to:
- Stage Five: The bone marrow becomes infected. At this point, the virus will stay with the cat for the rest of its life. In this phase, the virus replicates and is released four to seven days later in infected neutrophils, and sometimes lymphocytes, monocytes, and eosinophils (all white blood cells formed in the bone marrow).
- Stage Six: The cat's body is overwhelmed by infection and mucosal and glandular epithelial cells (tissue that forms a thin protective layer on exposed bodily surfaces and forms the lining of internal cavities, ducts, and organs) become infected. The virus replicates in epithelial tissues including salivary glands, oropharynx, stomach, esophagus, intestines, trachea, nasopharynx, renal tubules, bladder, pancreas, alveolar ducts, and sebaceous ducts from the muzzle.
Comparison with feline immunodeficiency virus
[edit]
FeLV and feline immunodeficiency virus (FIV) are sometimes mistaken for one another, though the viruses differ in many ways. Although they are both in the same retroviral subfamily (Orthoretrovirinae), they are classified in different genera (FeLV is a gamma-retrovirus and FIV is a lentivirus like HIV-1). Their shapes are quite different: FeLV is more circular while FIV is elongated. The two viruses are also quite different genetically, and their protein coats differ in size and composition. Although many of the diseases caused by FeLV and FIV are similar, the specific ways in which they are caused also differ. Also, while the feline leukemia virus may cause symptomatic illness in an infected cat, an FIV infected cat can remain completely asymptomatic its entire lifetime.[citation needed]
See also[edit]
https://en.wikipedia.org/wiki/Feline_leukemia_virus
Endogenous Retrovirus
See also[edit]
icon Viruses portal
Avian sarcoma leukosis virus (ASLV)
Endogenous viral element
ERV3
HERV-FRD
Horizontal gene transfer
Jaagsiekte sheep retrovirus (JSRV)
Koala retrovirus (KoRV)
Mouse mammary tumor virus (MMTV)
Murine leukemia virus (MLV) and xenotropic murine leukemia virus-related virus (XMRV)
Paleovirology
In mammals, intact env proteins called syncytins are responsible for the formation and function of syncytiotrophoblasts.[15] These multinucleated cells are mainly responsible for maintaining nutrient exchange and separating the fetus from the mother's immune system.[15] It has been suggested that the selection and fixation of these proteins for this function have played a critical role in the evolution of viviparity.[30]
https://en.wikipedia.org/wiki/Endogenous_retrovirus
https://en.wikipedia.org/wiki/Pospiviroidae
https://en.wikipedia.org/wiki/Avsunviroidae
https://en.wikipedia.org/wiki/Defective_interfering_particle
https://en.wikipedia.org/wiki/Fungal_prion
https://en.wikipedia.org/wiki/Mobilome
Self Replicating ORganic Structures; Genetic Transplantation Vectors at GMO genetically modified organism food crop etc.
https://en.wikipedia.org/wiki/Endogenous_retrovirus
https://en.wikipedia.org/wiki/Transpoviron
https://en.wikipedia.org/wiki/Extrachromosomal_DNA
Extrachromosomal circular DNA (eccDNA) are circular DNA found in human, plant and animal cells in addition to chromosomal DNA. eccDNA originate from chromosomal DNA and can be from 50 base pairs to several mega-base pairs in length and encode regulatory elements and several full genes.
eccDNA was first discovered in 1964 by Alix Bassel and Yasuo Hoota[1] in wheat nuclei and boar sperm.[2] Since then, eccDNA has been observed in almost all organisms from plants, yeast, C. elegans, frogs, mice, chicken, birds, and humans.[3][4][5][6][7][8][9] eccDNA molecules originate in normal cells and are a by product of programmed DNA recombination events; such as V(D)J recombination.[10][9] Moreover, eccDNA production seems to be cell-type specific in somatic cells.[9]
A subtype of eccDNA, such as ecDNA, ribosomal DNA locus (Extrachromosomal rDNA circle), and double minutes have been associated with genomic instability. Double minute ecDNAs are fragments of extrachromosomal DNA, which were originally observed in a large number of human tumors including breast, lung, ovary, colon, and most notably, neuroblastoma. They are a manifestation of gene amplification during the development of tumors, which give the cells selective advantages for growth and survival. Double minutes, like actual chromosomes, are composed of chromatinand replicate in the nucleus of the cell during cell division. Unlike typical chromosomes, they are composed of circular fragments of DNA, up to only a few million base pairs in size and contain no centromere or telomere.
See also[edit]
https://en.wikipedia.org/wiki/Extrachromosomal_circular_DNA#Role_of_ecDNA_in_cancer
Plasmodium is a genus of unicellular eukaryotes that are obligate parasites of vertebrates and insects. The life cycles of Plasmodium species involve development in a blood-feeding insect host which then injects parasites into a vertebrate host during a blood meal. Parasites grow within a vertebrate body tissue (often the liver) before entering the bloodstream to infect red blood cells. The ensuing destruction of host red blood cells can result in disease, called malaria. During this infection, some parasites are picked up by a blood-feeding insect (mosquitoes in majority cases), continuing the life cycle.[1]
https://en.wikipedia.org/wiki/Plasmodium
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently.
https://en.wikipedia.org/wiki/Plasmid
A phagemid or phasmid is a DNA-based cloning vector, which has both bacteriophage and plasmid properties.[1] These vectors carry, in addition to the origin of plasmid replication, an origin of replication derived from bacteriophage. Unlike commonly used plasmids, phagemid vectors differ by having the ability to be packaged into the capsid of a bacteriophage, due to their having a genetic sequence that signals for packaging. Phagemids are used in a variety of biotechnology applications; for example, they can be used in a molecular biology technique called "Phage Display".[2]
Filamentous phages retard bacterial growth but, contrasting with the lambda phage and the T7 phage, are not generally lytic. Helper phages are usually engineered to package less efficiently (via a defective phage origin of replication)[4] than the phagemid so that the resultant phage particles contain predominantly phagemid DNA. F1 Filamentous phage infection requires the presence of a pilus so only bacterial hosts containing the F-plasmid or its derivatives can be used to generate phage particles.
Prior to the development of cycle sequencing, phagemids were used to generate single stranded DNA template for sequencing purposes. Today phagemids are still useful for generating templates for site-directed mutagenesis. Detailed characterisation of the filamentous phage life cycle and structural features lead to the development of phage display technology, in which a range of peptides and proteins can be expressed as fusions to phage coat proteins and displayed on the viral surface. The displayed peptides and polypeptides are associated with the corresponding coding DNA within the phage particle and so this technique lends itself to the study of protein-protein interactions and other ligand/receptor combinations.
https://en.wikipedia.org/wiki/Phagemid
https://en.wikipedia.org/wiki/Phagemid
Dinodnavirus is a genus of viruses that infect dinoflagellates.[1] This genus belongs to the clade of nucleocytoplasmic large DNA viruses. The name is derived from 'dino' (dinoflagellate) and DNA (from its genome).
The only species in the genus is Heterocapsa circularisquama DNA virus 01.[2]
https://en.wikipedia.org/wiki/Dinodnavirus
Rhizidiovirus is a genus of viruses. Stramenopiles[1] (fungi and hyphochytridiomycota) serve as natural hosts. There is only one species in this genus: Rhizidiomyces virus.[2][3]
https://en.wikipedia.org/wiki/Rhizidiovirus
Plasmaviridae is a family of bacteria-infecting viruses. Acholeplasma species serve as natural hosts. There is one genus in the family, Plasmavirus, which contains one species: Acholeplasma virus L2.[1][2][3] All viruses known in this family have been isolated from species in the class Mollicutes.
This family is poorly studied and little is known about the diversity and biology of these viruses.
https://en.wikipedia.org/wiki/Plasmaviridae
A polydnavirus /pɒˈlɪdnəvaɪrəs/ (PDV) is a member of the family Polydnaviridae of insect viruses. There are two genera in the family: Bracovirus and Ichnovirus. Polydnaviruses form a symbiotic relationship with parasitoid wasps (ichnoviruses (IV) occur in ichneumonid wasps species and bracoviruses (BV) in braconid wasps), but these wasps are themselves parasitic on Lepidoptera (moths and butterflies).[1][2] Little or no sequence homology exists between BV and IV, suggesting that the two genera have been evolving independently for a long time.
https://en.wikipedia.org/wiki/Polydnavirus
Finnlakeviridae is a family of bacterial viruses that is not assigned to any higher taxonomic ranks. The family contains a single genus, Finnlakevirus, which contains a single species, Flavobacterium virus FLiP.[1][2] This virus was isolated in 2010, with its gram-negative host bacterium, from Lake Jyväsjärvi, a boreal freshwater habitat in Central Finland, and is the first described single-stranded DNA virus with an internal membrane.[2]
https://en.wikipedia.org/wiki/Finnlakeviridae
A nanobe[pronunciation?] is a tiny filamental structure first found in some rocks and sediments. Some scientists hypothesize that nanobes are the smallest form of life, 1/10 the size of the smallest known bacteria.[1]
No conclusive evidence exists that these structures are, or are not, living organisms, so their classification is controversial.
The 1996 discovery of nanobes was published in 1998[2] by Philippa Uwins et al.,[3] from the University of Queensland, Australia. They were found growing from rock samples (both full-diameter and sidewall cores) of Jurassic and Triassic sandstones, originally retrieved from an unspecified number of oil exploration wells off Australia's west coast. Depths of retrieval were between 3,400 metres (2.1 mi) and 5,100 metres (3.2 mi) below the sea bed. While Uwins et al. present assertions against it, they do not exclude the possibility that the nanobes are from a surface contaminant, not from the rock units cited.
The smallest are just 20 nanometers in diameter. Some researchers believe that these structures are crystal growths, but the staining of these structures with dyes that bind to DNA might indicate that they are living organisms.[4] They are similar to the structures found in ALH84001, a Mars meteorite found in the Antarctic. Nanobes are similar in size to nanobacteria, which are also structures that had been proposed to be extremely small living organisms. However, these two should not be confused. Nanobacteria were thought to be cellular organisms, while nanobes are hypothesized (by some) to be a previously unknown form of life or protocells.[citation needed]
https://en.wikipedia.org/wiki/Nanobe
Ultramicrobacteria are bacteria that are smaller than 0.1 μm3 under all growth conditions.[1][2][3] This term was coined in 1981, describing cocci in seawater that were less than 0.3 μm in diameter.[4] Ultramicrobacteria have also been recovered from soil and appear to be a mixture of Gram-positive, Gram-negative and cell-wall-lacking species.[5][2] Ultramicrobacteria possess a relatively high surface-area-to-volume ratio due to their small size, which aids in growth under oligotrophic (i.e. nutrient-poor) conditions.[2] The relatively small size of ultramicrobacteria also enables parasitism of larger organisms;[2] some ultramicrobacteria have been observed to be obligate or facultative parasites of various eukaryotes and prokaryotes.[1][2]One factor allowing ultramicrobacteria to achieve their small size seems to be genome minimization[1][2] such as in the case of the ultramicrobacterium P. ubique whose small 1.3 Mb genome is seemingly devoid of extraneous genetic elements like non-coding DNA, transposons, extrachromosomal elements etc.[2] However, genomic data from ultramicrobacteria is lacking[2] since the study of ultramicrobacteria, like many other prokaryotes, is hindered by difficulties in cultivating them.[3]
Ultramicrobacteria are commonly confused with ultramicrocells, the latter of which are the dormant, stress-resistant forms of larger cells that form under starvation conditions[1][2][7] (i.e. these larger cells downregulate their metabolism, stop growing and stabilize their DNA to create ultramicrocells that remain viable for years[1][8]) whereas the small size of ultramicrobacteria is not a starvation response and is consistent even under nutrient-rich conditions.[3]
The term "nanobacteria" is sometimes used synonymously with ultramicrobacteria in the scientific literature,[2] but ultramicrobacteria are distinct from the purported nanobacteria or "calcifying nanoparticles", which were proposed to be living organisms that were 0.1 μm in diameter.[9] These structures are now thought to be non-living,[10] and likely precipitated particles of inorganic material.[11][12]
See also[edit]
- L-form bacteria
- Mycoplasma – smallest known bacteria (300 nm)
- Nanoarchaeum – smallest known archaeum (400 nm)
- Nanobacteria – possible lifeforms smaller than bacteria (<200 nm)
- Nanobe – possible smallest lifeforms (20 nm)
- Pandoravirus – one of the largest known viruses (1000 nm)
- Parvovirus – smallest known viruses (18–28 nm)
- Pithovirus – largest known virus (1500 nm)
- Prion – smallest known infectious agent (≈10 nm)
- ND5 and MY14T – two aerobic, Gram-negative, rod-shaped bacteria[13]
https://en.wikipedia.org/wiki/Ultramicrobacteria
L-form bacteria, also known as L-phase bacteria, L-phase variants, and cell wall-deficient (CWD) bacteria, are strains of bacteria that lack cell walls.[1] They were first isolated in 1935 by Emmy Klieneberger-Nobel, who named them "L-forms" after the Lister Institute in London where she was working.[2]
Two types of L-forms are distinguished: unstable L-forms, spheroplasts that are capable of dividing, but can revert to the original morphology, and stable L-forms, L-forms that are unable to revert to the original bacteria.
Some parasitic species of bacteria, such as mycoplasma, also lack a cell wall,[3] but these are not considered L-forms since they are not derived from bacteria that normally have cell walls.[4]
https://en.wikipedia.org/wiki/L-form_bacteria
Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants and humans. As a member of the genusBacillus, B. subtilis is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. B. subtilis has historically been classified as an obligate aerobe, though evidence exists that it is a facultative anaerobe. B. subtilis is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies.[3]
| Bacillus subtilis | |
|---|---|
 | |
| TEM micrograph of a B. subtilis cell in cross-section (scale bar = 200 nm) |
https://en.wikipedia.org/wiki/Bacillus_subtilis
Methotrexate (MTX), formerly known as amethopterin, is a chemotherapy agent and immune-system suppressant.[4] It is used to treat cancer, autoimmune diseases, and ectopic pregnancyand for medical abortions.[4] Types of cancers it is used for include breast cancer, leukemia, lung cancer, lymphoma, gestational trophoblastic disease, and osteosarcoma.[4] Types of autoimmune diseases it is used for include psoriasis, rheumatoid arthritis, and Crohn's disease.[4] It can be given by mouth or by injection.[4]
https://en.wikipedia.org/wiki/Methotrexate
L-forms can be generated in the laboratory from many bacterial species that usually have cell walls, such as Bacillus subtilis or Escherichia coli. This is done by inhibiting peptidoglycan synthesis with antibiotics or treating the cells with lysozyme, an enzyme that digests cell walls. The L-forms are generated in a culture medium that is the same osmolarity as the bacterial cytosol (an isotonic solution), which prevents cell lysis by osmotic shock.[2] L-form strains can be unstable, tending to revert to the normal form of the bacteria by regrowing a cell wall, but this can be prevented by long-term culture of the cells under the same conditions that were used to produce them – letting the wall-disabling mutations to accumulate by genetic drift.[6]
Some studies have identified mutations that occur, as these strains are derived from normal bacteria.[1][2]One such point mutation D92E is in an enzyme yqiD/ispA (P54383) involved in the mevalonate pathwayof lipid metabolism that increased the frequency of L-form formation 1,000-fold.[1] The reason for this effect is not known, but it is presumed that the increase is related to this enzyme's role in making a lipid important in peptidoglycan synthesis.
Another methodology of induction relies on nanotechnology and landscape ecology. Microfluidics devices can be built in order to challenge peptidoglycan synthesis by extreme spatial confinement. After biological dispersal through a constricted (sub-micrometre scale) biological corridor connecting adjacent micro habitat patches, L-form-like cells can be derived[7] using a microfluifics-based (synthetic) ecosystem implementing an adaptive landscape[8] selecting for shape-shifting phenotypes similar to L-forms.
https://en.wikipedia.org/wiki/L-form_bacteria
Significance and applications
[edit]
Some publications have suggested that L-form bacteria might cause diseases in humans,[9] and other animals[10] but, as the evidence that links these organisms to disease is fragmentary and frequently contradictory, this hypothesis remains controversial.[11][12] The two extreme viewpoints on this question are that L-form bacteria are either laboratory curiosities of no clinical significance or important but unappreciated causes of disease.[4]Research on L-form bacteria is continuing. For example, L-form organisms have been observed in mouse lungs after experimental inoculation with Nocardia caviae,[13][14] and a recent study suggested that these organisms may infect immunosuppressed patients having undergone bone marrow transplants.[15] The formation of strains of bacteria lacking cell walls has also been proposed to be important in the acquisition of bacterial antibiotic resistance.[16][17]
L-form bacteria may be useful in research on early forms of life, and in biotechnology. These strains are being examined for possible uses in biotechnology as host strains for recombinant protein production.[18][19][20] Here, the absence of a cell wall can allow production of large amounts of secreted proteins that would otherwise accumulate in the periplasmic space of bacteria.[21][22]
See also[edit]
- Mycoplasmataceae—lack peptidoglycan but supplement their membranes with sterols for stability.
- Protoplast
- Spheroplast
- Ultramicrobacteria
Mycoplasmataceae is a family of bacteria[1] in the order Mycoplasmatales. This family consists of the genera Mycoplasma and Ureaplasma.
In 1967, the order Mycoplasmatales was incorporated into the class Mollicutes.[2] Many species are sexually transmitted and cause pelvic inflammatory disease.[3]
Mycoplasma refers to a genus of bacteria that lack a cell wall and possess a three-layered cellular membrane.[4] They can be parasitic or saprotrophic. Several species are sexually transmitted and pathogenic in humans. Others are found on cats, dogs, and barnyard fowl.
Ureaplasma spp. as human pathogens[edit]
Both Ureaplasma urealyticum and Ureaplasma parvum have been identified as important human pathogens, causing infection in the urogenital tract and, rarely, at distal sites.[5][6][7] Their role in neonatal disease and adverse pregnancy outcomes has been well established, and semantic classifications are changing to reflect the nature of the detrimental outcomes these infections are associated with.[8] In the 2010s, Mycoplasma genitalium has been re-classified as an STI, and it is likely that with more research, Ureaplasma spp. will follow this trend.[9] Similar to other pathogens such as Chlamydia trachomatis, infection with Ureaplasma spp. is associated with adverse fertility outcomes in both men and women.[10][11][12][13][14]Both cause non-gonococcal urethritis. Ureaplasma spp. were implicated in conditions such as prostatitis and chronic pelvic pain syndrome as early as the 1980s.[15][16][17] Research in women has lagged several decades behind, but it is now becoming more clear how Ureaplasma spp. contribute to etiologies such as interstitial cystitis/painful bladder syndrome.[18][19][20] Ureaplasma spp. are associated with alterations in host environment that increase susceptibility to other infections such as bacterial vaginosis and vaginal candidiasis.[21][22] Ureaplasma spp. can cause reactive arthritis as well as directly infect the synovium.[23][7] Some case studies have suggested a causative role in complex regional pain syndrome/reflex sympathetic dystrophy syndrome.[24]
| Mycoplasmataceae | |
|---|---|
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | Mycoplasmatales |
| Family: | Mycoplasmataceae Freundt 1955 |
| Genera | |
Candidatus Hepatoplasma | |
https://en.wikipedia.org/wiki/Mycoplasmataceae
Mollicutes is a class of bacteria[2] distinguished by the absence of a cell wall. The word "Mollicutes" is derived from the Latin mollis (meaning "soft" or "pliable"), and cutis (meaning "skin"). Individuals are very small, typically only 0.2–0.3 μm (200-300 nm) in size and have a very small genome size. They vary in form, although most have sterols that make the cell membrane somewhat more rigid. Many are able to move about through gliding, but members of the genus Spiroplasma are helical and move by twisting. The best-known genus in the Mollicutes is Mycoplasma.
Mollicutes are parasites of various animals and plants, living on or in the host's cells. Many cause diseases in humans, attaching to cells in the respiratory or urogenital tracts, particularly species of Mycoplasma and Ureaplasma. Phytoplasma and Spiroplasma are plant pathogens associated with insect vectors.
Whereas formerly the trivial name "mycoplasma" has commonly denoted any member of the class Mollicutes, it now refers exclusively to a member of the genus Mycoplasma.
| Mollicutes | |
|---|---|
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Tenericutes |
| Class: | Mollicutes Edward and Freundt 1967[1] |
| Orders | |
Acholeplasmatales | |
https://en.wikipedia.org/wiki/Mollicutes
Tenericutes (tener cutis: soft skin) is a phylum of bacteria that contains the class Mollicutes. The name was validated in 1984 as a new division (phylum).[3][4][5] Notable genera include Mycoplasma, Spiroplasma, Ureaplasma, and Phytoplasma.
https://en.wikipedia.org/wiki/Tenericutes
At first, all members of the class Mollicutes were generally named "mycoplasma" or pleuropneumonia-like organism (PPLO). Mollicutes other than some members of genus Mycoplasma were still unidentified. The first species of Mycoplasma/Mollicutes, that could be isolated was Mycoplasma mycoides. This bacterium was cultivated by Nocard and Roux in 1898.[4]
In 1962, R.G.E. Murray proposed to divide the kingdom Bacteria into three divisions (= phyla) on the basis of the cell wall types:
- Gram-negative Gracilicutes, with a thin cell wall and little peptidoglycan;
- Gram-positive "Firmacutes", with a thicker cell wall and more peptidoglycan (the name was later changed in "Firmicutes"), and
- the "Mollicutes", without a cell wall.[6]
https://en.wikipedia.org/wiki/Mollicutes
https://en.wikipedia.org/wiki/Planctomycetes
https://en.wikipedia.org/wiki/Verrucomicrobia
https://en.wikipedia.org/wiki/Chlamydiae
https://en.wikipedia.org/wiki/Planctobacteria
https://en.wikipedia.org/wiki/Aquificae
https://en.wikipedia.org/wiki/Phagemid
https://en.wikipedia.org/wiki/Spumaretrovirinae
https://en.wikipedia.org/wiki/Bovine_leukemia_virus
https://en.wikipedia.org/wiki/Rhizidiovirus
https://en.wikipedia.org/wiki/Plasmaviridae
https://en.wikipedia.org/wiki/Polydnavirus
https://en.wikipedia.org/wiki/Finnlakeviridae
https://en.wikipedia.org/wiki/Mollicutes
https://en.wikipedia.org/wiki/Mycoplasmataceae
https://en.wikipedia.org/wiki/Mollicutes
https://en.wikipedia.org/wiki/Methotrexate
https://en.wikipedia.org/wiki/Treponema_pallidum
https://en.wikipedia.org/wiki/Myxococcus
https://en.wikipedia.org/wiki/Phagemid
https://en.wikipedia.org/wiki/Bacillus_subtilis
https://en.wikipedia.org/wiki/Gammaretrovirus
https://en.wikipedia.org/wiki/Woolly_monkey_sarcoma_virus
https://en.wikipedia.org/wiki/Reticuloendotheliosis_virus
https://en.wikipedia.org/wiki/Chick_syncytial_virus
https://en.wikipedia.org/wiki/Gammaretrovirus
https://en.wikipedia.org/wiki/Spumaretrovirinae
https://en.wikipedia.org/wiki/Progressive_vaccinia
https://en.wikipedia.org/wiki/Generalized_vaccinia
https://en.wikipedia.org/wiki/Vaccinia
https://en.wikipedia.org/wiki/Palmoplantar_keratoderma
https://en.wikipedia.org/wiki/Dystrophic_calcification
https://en.wikipedia.org/wiki/Calciphylaxis
https://en.wikipedia.org/wiki/NF-κB
https://en.wikipedia.org/wiki/Endogenous_retrovirus#Human_endogenous_retroviruses
https://en.wikipedia.org/wiki/Gammaretrovirus
https://en.wikipedia.org/wiki/Spumaretrovirinae
https://en.wikipedia.org/wiki/Bovine_leukemia_virus
https://en.wikipedia.org/wiki/Phagemid
https://en.wikipedia.org/wiki/Mycoplasmataceae
https://en.wikipedia.org/wiki/Mycobacterium_tuberculosis
https://en.wikipedia.org/wiki/Caseous_necrosis
https://en.wikipedia.org/wiki/Necrosis
https://en.wikipedia.org/wiki/Gas_gangrene
https://en.wikipedia.org/wiki/Syphilis
https://en.wikipedia.org/wiki/HIV
https://en.wikipedia.org/wiki/Pyroptosis
https://en.wikipedia.org/wiki/Immunosuppressive_drug
https://en.wikipedia.org/wiki/Human_foamy_virus
https://en.wikipedia.org/wiki/House_dust_mite
https://en.wikipedia.org/wiki/Crustacean
https://en.wikipedia.org/wiki/Parasitism
https://en.wikipedia.org/wiki/Neurosarcoidosis
https://en.wikipedia.org/wiki/Timeline_of_human_vaccines
https://en.wikipedia.org/wiki/Neurocysticercosis
https://en.wikipedia.org/wiki/Smallpox
https://en.wikipedia.org/wiki/Measles
https://en.wikipedia.org/wiki/Malaria
https://microbewiki.kenyon.edu/index.php/Granulosis_Virus
https://en.wikipedia.org/wiki/Rabies
https://en.wikipedia.org/wiki/Ricketts
https://en.wikipedia.org/wiki/Leishmania
https://en.wikipedia.org/wiki/Granulation
https://en.wikipedia.org/wiki/Allergy
https://en.wikipedia.org/wiki/Rhinitis
https://en.wikipedia.org/wiki/Rash
https://en.wikipedia.org/wiki/Chickenpox
https://en.wikipedia.org/wiki/Eczema_vaccinatum
https://en.wikipedia.org/wiki/Lupus_erythematosus
https://en.wikipedia.org/wiki/Chromosome_21
https://en.wikipedia.org/wiki/Calciphylaxis
https://en.wikipedia.org/wiki/Dystrophic_calcification
https://en.wikipedia.org/wiki/NF-κB
https://en.wikipedia.org/wiki/Endogenous_retrovirus#Human_endogenous_retroviruses
https://en.wikipedia.org/wiki/Gammaretrovirus
https://en.wikipedia.org/wiki/NK-92
https://en.wikipedia.org/wiki/Rhizomucor_pusillus
https://en.wikipedia.org/wiki/Human_T-lymphotropic_virus
https://en.wikipedia.org/wiki/Human_T-lymphotropic_virus_1
https://en.wikipedia.org/wiki/Feline_leukemia_virus
https://en.wikipedia.org/wiki/Tumor_lysis_syndrome
https://en.wikipedia.org/wiki/Tumor_necrosis_factor
https://en.wikipedia.org/wiki/Rhabdomyolysis
https://en.wikipedia.org/wiki/Caprine_arthritis_encephalitis_virus
https://en.wikipedia.org/wiki/Jaagsiekte_sheep_retrovirus
https://en.wikipedia.org/wiki/Visna-maedi_virus
https://en.wikipedia.org/wiki/Bacillus_thuringiensis
https://en.wikipedia.org/wiki/Lentivirus
https://en.wikipedia.org/wiki/Human_T-lymphotropic_virus
https://en.wikipedia.org/wiki/Simian_immunodeficiency_virus
https://en.wikipedia.org/wiki/Treponema_pallidum
https://seer.cancer.gov/seertools/hemelymph/51f6cf57e3e27c3994bd5363/?q=cytoid#
https://seer.cancer.gov/seertools/hemelymph/532b32a0e4b0626b1926e990/
https://en.wikipedia.org/wiki/Myxococcus
https://en.wikipedia.org/wiki/Methotrexate
https://en.wikipedia.org/wiki/Pseudomonas_aeruginosa
https://en.wikipedia.org/wiki/Leukemia
https://en.wikipedia.org/wiki/Provirus
https://en.wikipedia.org/wiki/Bacillus_coagulans
https://en.wikipedia.org/wiki/Monckeberg%27s_arteriosclerosis
https://en.wikipedia.org/wiki/NF-κB
https://en.wikipedia.org/wiki/Dystrophic_calcification
https://en.wikipedia.org/wiki/Generalized_vaccinia
https://en.wikipedia.org/wiki/Progressive_vaccinia
https://en.wikipedia.org/wiki/Wart
https://en.wikipedia.org/wiki/Eczema_vaccinatum
https://en.wikipedia.org/wiki/Category:Bacteria_described_in_1915
https://en.wikipedia.org/wiki/Calciphylaxis
https://en.wikipedia.org/wiki/NF-κB
https://en.wikipedia.org/wiki/Dystrophic_calcification
https://en.wikipedia.org/wiki/Human_foamy_virus
https://en.wikipedia.org/wiki/Avian_sarcoma_leukosis_virus
https://en.wikipedia.org/wiki/Allergic_rhinitis
https://en.wikipedia.org/wiki/Bacillus_subtilis
https://en.wikipedia.org/wiki/Bacillus_atrophaeus
https://en.wikipedia.org/wiki/Rotavirus
https://en.wikipedia.org/wiki/Shigellosis
https://en.wikipedia.org/wiki/Pollen
https://en.wikipedia.org/wiki/Microsporangia
https://en.wikipedia.org/wiki/Common_cold
https://en.wikipedia.org/wiki/Bacillus_subtilis
https://en.wikipedia.org/wiki/Bacillus_subtilis
https://en.wikipedia.org/wiki/Bacillus_atrophaeus
https://en.wikipedia.org/wiki/Radiotrophic_fungus
https://en.wikipedia.org/wiki/Psammophile
https://en.wikipedia.org/wiki/Psychrophile
https://en.wikipedia.org/wiki/Piezophile
https://en.wikipedia.org/wiki/Osmophile
https://en.wikipedia.org/wiki/Lipophilic_bacteria
https://en.wikipedia.org/wiki/Cytokinesis
https://en.wikipedia.org/wiki/Cytokine
https://en.wikipedia.org/wiki/Methylation
https://en.wikipedia.org/wiki/Category:Epigenetics
https://en.wikipedia.org/wiki/Epigenetic_clock
https://en.wikipedia.org/wiki/Category:Papillomavirus-associated_diseases
https://en.wikipedia.org/wiki/Molluscum_contagiosum_virus
https://en.wikipedia.org/wiki/Basal-cell_carcinoma
https://en.wikipedia.org/wiki/Herpesviridae
above. poor unfortunate souls ursula
NF-κB is found in almost all animal cell types and is involved in cellular responses to stimuli such as stress, cytokines, free radicals, heavy metals, ultraviolet irradiation, oxidized LDL, and bacterial or viral antigens.[2][3][4][6][7] NF-κB plays a key role in regulating the immune response to infection. Incorrect regulation of NF-κB has been linked to cancer, inflammatory and autoimmune diseases, septic shock, viral infection, and improper immune development. NF-κB has also been implicated in processes of synaptic plasticity and memory.[8][9][10][11][12][13]
https://en.wikipedia.org/wiki/NF-κB
Human parainfluenza viruses (HPIVs) are the viruses that cause human parainfluenza. HPIVs are a paraphyletic group of four distinct single-stranded RNA viruses belonging to the Paramyxoviridae family. These viruses are closely associated with both human and veterinary disease.[2] Virions are approximately 150–250 nm in size and contain negative sense RNA with a genome encompassing about 15,000 nucleotides.[3]
The viruses can be detected via cell culture, immunofluorescent microscopy, and PCR.[4] HPIVs remain the second main cause of hospitalisation in children under 5 years of age suffering from a respiratory illness (only Human orthopneumovirus, or Respiratory syncytial virus (RSV), causes more respiratory hospitalisations for this age group).[5]
https://en.wikipedia.org/wiki/Human_parainfluenza_viruses
The first HPIV was discovered in the late 1950s. The taxonomic division is broadly based on antigenic and genetic characteristics, forming four major serotypes or clades, which today are considered distinct viruses.[6]These include:
| Virus | GenBank acronym | NCBI taxonomy | Notes |
|---|---|---|---|
| Human parainfluenza virus type 1 | HPIV-1 | 12730 | Most common cause of croup |
| Human parainfluenza virus type 2 | HPIV-2 | 11212 | Causes croup and other upper and lower respiratory tract illnesses |
| Human parainfluenza virus type 3 | HPIV-3 | 11216 | Associated with bronchiolitis and pneumonia |
| Human parainfluenza virus type 4 | HPIV-4 | 11203 | Includes subtypes 4a and 4b |
HPIVs belong to two genera: Respirovirus (HPIV-1 & HPIV-3) and Rubulavirus (HPIV-2 & HPIV-4).[3]
https://en.wikipedia.org/wiki/Human_parainfluenza_viruses
Pneumoviridae (from Greek pneumo-, lung, -viridae, virus from Latin, poison, slimy liquid)[2][3] is a family of negative-strand RNA viruses in the order Mononegavirales.[1][4] Humans, cattle, and rodents serve as natural hosts.[5] Respiratory tract infections are associated with member viruses such as human respiratory syncytial virus. There are five species in the family which are divided between the genera Metapneumovirus and Orthopneumovirus. The family used to be considered as a sub-family of Paramyxoviridae, but has been reclassified as of 2016.[1]
| Genus | Species | Virus (Abbreviation) |
| Metapneumovirus | Avian metapneumovirus | avian metapneumovirus (AMPV) |
| Human metapneumovirus | human metapneumovirus (HMPV) | |
| Orthopneumovirus | Bovine orthopneumovirus | bovine respiratory syncytial virus (BRSV) |
| Human orthopneumovirus | human respiratory syncytial virus A2 (HRSV-A2) | |
| human respiratory syncytial virus B1 (HRSV-B1) | ||
| Murine orthopneumovirus | murine pneumonia virus (MPV) |
https://en.wikipedia.org/wiki/Pneumoviridae
https://en.wikipedia.org/wiki/Human_parainfluenza_viruses
https://en.wikipedia.org/wiki/Crimean–Congo_hemorrhagic_fever
Thogotovirus is a genus of enveloped RNA viruses, one of seven genera in the virus family Orthomyxoviridae. Their single-stranded, negative-sense RNA genome has six or seven segments. Thogotoviruses are distinguished from most other orthomyxoviruses[3] by being arboviruses – viruses that are transmitted by arthropods, in this case usually ticks. Thogotoviruses can replicate in both tick cells and vertebrate cells; one subtype has also been isolated from mosquitoes. A consequence of being transmitted by blood-sucking vectors is that the virus must spread systemically in the vertebrate host – unlike influenza viruses, which are transmitted by respiratory droplets and are usually confined to the respiratory system.[4]
The genus contains the species Thogoto thogotovirus and Dhori virus (DHOV), and the latter's subtype Batken virus, as well as the species or strains Araguari virus, Aransas Bay virus (ABV), Bourbon virus, Jos virus (JOSV) and Upolu virus (UPOV), which have yet to be confirmed by the International Committee on Taxonomy of Viruses (ICTV). A wide range of mammals are infected by members of the genus; some types also infect birds. THOV causes disease in livestock. THOV, DHOV and Bourbon virus can infect humans, and have occasionally been associated with human disease.
https://en.wikipedia.org/wiki/Thogotovirus
Arbovirus is an informal name for any virus that is transmitted by arthropod vectors. The term arbovirus is a portmanteau word (arthropod-borne virus).[1] Tibovirus (tick-borne virus) is sometimes used to more specifically describe viruses transmitted by ticks, a superorder within the arthropods.[2] Arboviruses can affect both animals (including humans) and plants. In humans, symptoms of arbovirus infection generally occur 3–15 days after exposure to the virus and last three or four days. The most common clinical features of infection are fever, headache, and malaise, but encephalitis and viral hemorrhagic fever may also occur.[3]
https://en.wikipedia.org/wiki/Arbovirus
Viral hemorrhagic fevers (VHFs) are a diverse group of animal and human illnesses in which fever and hemorrhage are caused by a viral infection. VHFs may be caused by five distinct families of RNA viruses: the families Filoviridae, Flaviviridae, Rhabdoviridae, and several member families of the Bunyavirales order such as Arenaviridae, and Hantaviridae. All types of VHF are characterized by fever and bleeding disorders and all can progress to high fever, shock and death in many cases. Some of the VHF agents cause relatively mild illnesses, such as the Scandinavian nephropathia epidemica (a hantavirus), while others, such as Ebola virus, can cause severe, life-threatening disease.
https://en.wikipedia.org/wiki/Viral_hemorrhagic_fever
The human immunodeficiency viruses (HIV) are two species of Lentivirus (a subgroup of retrovirus) that infect humans. Over time, they cause acquired immunodeficiency syndrome (AIDS),[1][2] a condition in which progressive failure of the immune system allows life-threatening opportunistic infections and cancers to thrive.[3] Without treatment, average survival time after infection with HIV is estimated to be 9 to 11 years, depending on the HIV subtype.[4] In most cases, HIV is a sexually transmitted infection and occurs by contact with or transfer of blood, pre-ejaculate, semen, and vaginal fluids. Research has shown (for both same-sex and opposite-sex couples) that HIV is untransmittable through condomless sexual intercourse if the HIV-positive partner has a consistently undetectable viral load.[5][6] Non-sexual transmission can occur from an infected mother to her infant during pregnancy, during childbirth by exposure to her blood or vaginal fluid, and through breast milk.[7][8][9][10] Within these bodily fluids, HIV is present as both free virus particles and virus within infected immune cells.
HIV infects vital cells in the human immune system, such as helper T cells (specifically CD4+ T cells), macrophages, and dendritic cells.[11] HIV infection leads to low levels of CD4+ T cells through a number of mechanisms, including pyroptosis of abortively infected T cells,[12] apoptosis of uninfected bystander cells,[13]direct viral killing of infected cells, and killing of infected CD4+ T cells by CD8+ cytotoxic lymphocytes that recognize infected cells.[14] When CD4+ T cell numbers decline below a critical level, cell-mediated immunityis lost, and the body becomes progressively more susceptible to opportunistic infections, leading to the development of AIDS.
https://en.wikipedia.org/wiki/HIV
Disenabled
- WNT3A pseudogene, associated with the growth of a tail[1]
- NCF1C pseudogene, associated with a type of white blood cell.[2] It makes part of the neutrophil NADPH oxidase enzyme, which makes superoxide anion.[3]
- GULO pseudogene, associated with the production of Vitamin C
- IRGM pseudogene, associated with the immune system[4]
- hHaA pseudogene, associated with fur-like body hair:[5] see hypertrichosis
- DEFT1P pseudogene, associated with the immune system[6]
- Urate oxidase pseudogene, associated with the processing of uric acid
- Photolyase pseudogene, associated with repairing DNA damaged by UV radiation
- Photolyase is no longer encoded for despite obvious advantages.[7] Instead, this gene is mutated to encode for cryptochromes.
Human Genome
By chromosome[edit]
Below is a list of articles on human chromosomes, each of which contains an incomplete list of genes located on that chromosome.
- Chromosome 1 (human)
- Chromosome 2 (human)
- Chromosome 3 (human)
- Chromosome 4 (human)
- Chromosome 5 (human)
- Chromosome 6 (human)
- Chromosome 7 (human)
- Chromosome 8 (human)
- Chromosome 9 (human)
- Chromosome 10 (human)
- Chromosome 11 (human)
- Chromosome 12 (human)
- Chromosome 13 (human)
- Chromosome 14 (human)
- Chromosome 15 (human)
- Chromosome 16 (human)
- Chromosome 17 (human)
- Chromosome 18 (human)
- Chromosome 19 (human)
- Chromosome 20 (human)
- Chromosome 21 (human)
- Chromosome 22 (human)
- Chromosome X (human)
- Chromosome Y (human)
Transcription factors[edit]
This is a list of 1639 genes which encode proteins that are known or expected to function as human transcription factors.
See also[edit]
References[edit]
External links[edit]
- iHOP-Protein Information Database
- NextBio-Life Science Search Engine
- Entrez-Cross Database Query Search System
- TranscriptomeBrowser
- Cancers: Rearrangements (translocations) of genetic material between chromosome 21 and other chromosomes have been associated with several types of cancer. For example, acute lymphoblastic leukemia (a type of blood cancer most often diagnosed in childhood) has been associated with a translocation between chromosomes 12 and 21. Another form of leukemia, acute myeloid leukemia, has been associated with a translocation between chromosomes 8 and 21.
- Duplication in Amyloid precursor protein (APP) locus (duplicated segment varies in length but includes APP) on Chromosome 21 was found to cause early onset familial Alzheimer's disease in a French family set (Rovelet-Lecrux et al.) and a Dutch family set.[14] Compared to Alzheimer's caused by missense mutations in APP, the frequency of the Alzheimer's caused by APP duplications is significant. All patients that have an extra copy of APP gene due to the locus duplication show Alzheimer's with severe cerebral amyloid angiopathy.
G-bands of human chromosome 21 in resolution 850 bphs[4] Chr. Arm[20] Band[21] ISCN
start[22]ISCN
stop[22]Basepair
startBasepair
stopStain[23] Density 21 p 13 0 311 1 3,100,000 gvar 21 p 12 311 683 3,100,001 7,000,000 stalk 21 p 11.2 683 1056 7,000,001 10,900,000 gvar 21 p 11.1 1056 1274 10,900,001 12,000,000 acen 21 q 11.1 1274 1367 12,000,001 13,000,000 acen 21 q 11.2 1367 1584 13,000,001 15,000,000 gneg 21 q 21.1 1584 2019 15,000,001 22,600,000 gpos 100 21 q 21.2 2019 2144 22,600,001 25,500,000 gneg 21 q 21.3 2144 2330 25,500,001 30,200,000 gpos 75 21 q 22.11 2330 2485 30,200,001 34,400,000 gneg 21 q 22.12 2485 2610 34,400,001 36,400,000 gpos 50 21 q 22.13 2610 2703 36,400,001 38,300,000 gneg 21 q 22.2 2703 2858 38,300,001 41,200,000 gpos 50 21 q 22.3 2858 3200 41,200,001 46,709,983 gneg
Number of genes[edit]
The following are some of the gene count estimates of human chromosome 1. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[7]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 1,961 | — | — | [2] | 2016-09-08 |
| HGNC | 1,993 | 707 | 1,113 | [8] | 2017-05-12 |
| Ensembl | 2,044 | 1,924 | 1,223 | [9] | 2017-03-29 |
| UniProt | 2,064 | — | — | [10] | 2018-02-28 |
| NCBI | 2,093 | 1,790 | 1,426 | [11][12][13] | 2017-05-19 |
Gene list[edit]
The following is a partial list of genes on human chromosome 1. For complete list, see the link in the infobox on the right.
- DENN1B: hypothesized to be related to asthma
- LOC100132287: uncharacterized protein
p-arm[edit]
Partial list of the genes located on p-arm (short arm) of human chromosome 1:
- AADACL3: Arylacetamide deacetylase-like 3
- AADACL4: Arylacetamide deacetylase-like 4
- ACADM: acyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain
- ACTL8: Actin-like 8
- ADGRL2 (1p31.1): adhesion G protein-coupled receptor L2
- ADPRHL2: Poly(ADP-ribose) glycohydrolase ARH3
- AMPD2: encoding enzyme AMP deaminase 2
- ARID1A (1p36)
- ATXN7L2: Ataxin 7-like 2
- AZIN2: encoding enzyme Antizyme inhibitor 2 (AzI2) also known as arginine decarboxylase (ADC)
- BCAS2: Breast carcinoma amplified sequence 2
- BCL10 (1p22)
- BCL2L15 (1p13)
- LRIF1: encoding protein Ligand-dependent nuclear receptor-interacting factor 1
- C1orf109: chromosome 1 open reading frame 109
- CZIB: chromosome 1 open reading frame 123
- CACHD1 encoding protein Cache domain containing 1
- CAMTA1 (1p36)
- CASP9 (1p36)
- CASZ1 (1p36): Castor zinc finger 1
- CSDE1: Cold shock domain containing E1
- CHD5 (1p36)
- CLIC4 (1p36)
- CLSPN (1p34)
- CMPK: UMP-CMP kinase
- COL16A1 (1p35)
- COL11A1: collagen, type XI, alpha 1
- CPT2: carnitine palmitoyltransferase II
- CRYZ: Crystallin zeta
- CYP4B1 (1p33)
- CYR61 (1p22)
- DBT: dihydrolipoamide branched chain transacylase E2
- DCLRE1B: DNA cross-link repair 1B
- DEPDC1 encoding protein DEP domain containing 1
- DIRAS3 (1p31): DIRAS family, GTP-binding RAS-like 3
- DPH5: Diphthine synthase
- DVL1 (1p36)
- ENO1 (1p36)
- EPHA2 (1p36)
- EPS15 (1p32)
- ESPN: espin (autosomal recessive deafness 36)
- EVI5: ecotropic viral integration site 5
- EXTL1: exostosin like glycosyltransferase 1
- EXTL2: exostosin like glycosyltransferase 2
- FAM46B: family with sequence similarity 46, member B
- FAM46C: family with sequence similarity 46, member C
- FAM76A: family with sequence similarity 76, member A
- FBXO2: F-box protein 2
- FNBP1L encoding protein Formin-binding protein 1-like
- FPGT: Fucose-1-phosphate guanylyltransferase
- FUBP1 (1p31)
- GALE: UDP-galactose-4-epimerase
- GADD45A (1p31)
- GBP1 (1p22)
- GBP2: guanylate binding protein 2
- GBP5 encoding protein Guanylate binding protein 5
- GJB3: gap junction protein, beta 3, 31kDa (connexin 31)
- GLMN (1p22)
- GNL2: G protein nucleolar 2
- GSTM1 (1p13)
- HDAC1 (1p35)
- HES2: Hes family bHLH transcription factor 2
- HES3: Hes family bHLH transcription factor 3
- HMGCL: 3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase (hydroxymethylglutaricaciduria)
- HAO2 encoding protein Hydroxyacid oxidase 2
- HMGCS2: 3-hydroxy-3-methylglutaryl-CoA synthase 2
- HP1BP3: Heterochromatin protein 1, binding protein 3
- IFI6: Interferon alpha-inducible protein 6
- IL22RA1 (1p36)
- INTS11: Integrator complex subunit 11
- JAK1 (1p31)
- JUN (1p32)
- KCNQ4: potassium voltage-gated channel, KQT-like subfamily, member 4
- KIF1B: kinesin family member 1B
- L1TD1: LINE-1 type transposase domain containing 1
- LCK (1p35)
- LRRC39: Leucine-rich repeat-containing protein 39
- LRRC40: Leucine-rich repeat-containing protein 40
- LRRC41: Leucine-rich repeat-containing protein 41
- LRRC8D: Leucine-rich repeat-containing protein 8D
- MAN1A2: Mannosyl-oligosaccharide 1,2-alpha-mannosidase IB
- MEAF6: MYST/ESA1 associated factor 6
- MECR: Trans-2-enoyl-CoA reductase, mitochondrial
- MFAP2: Microfibrillar-associated protein 2
- MIB2 (1p36)
- MIER1 (1p31)
- MFN2: mitofusin 2
- MFSD2: Major facilitator superfamily domain containing 2A
- MIR6079: microRNA 6079
- MMEL1: Membrane metallo-endopeptidase-like 1
- MTFR1L: mitochondrial fission regulator 1 like
- MTHFR (1p36): 5,10-methylenetetrahydrofolate reductase (NADPH)
- MUL1: Mitochondrial E3 ubiquitin protein ligase 1
- MUTYH (1p34): mutY homolog (E. coli)
- NBPF3: Neuroblastoma breakpoint family member 3
- NGF: Nerve Growth Factor
- NOL9: Nucleolar protein 9
- NRAS (1p13)
- NOTCH2 (1p12)
- OLFML3: Olfactomedin-like 3
- OMA1: Metalloendopeptidase OMA1, mitochondrial
- OVGP1: Oviductal glycoprotein 1
- PARK7 (1p36): Parkinson disease (autosomal recessive, early onset) 7
- PINK1: PTEN induced putative kinase 1
- PLOD1: procollagen-lysine 1, 2-oxoglutarate 5-dioxygenase 1
- PRMT6: Protein arginine methyltransferase 6
- PSRC1: Proline/serine-rich coiled-coil protein 1
- RAD54L: RAD54-like
- RAP1A (1p13)
- RBM15 (1p13)
- RCC2: Regulator of chromosome condensation 2
- REG4 (1p12)
- RHBDL2: Rhomboid like 2
- RHOC (1p13)
- RLF: rearranged L-myc fusion
- RNF11 (1p32)
- RNF220: RING finger protein 220
- RPA2 (1p35)
- RSPO1 (1p34)
- S100A1 (1q21)
- SDC3: Syndecan-3
- SDHB (1p36)
- SFPQ (1p34)
- SGIP1: SH3 domain GRB2-like protein 3-interaction protein 1
- SH3BGRL3: SH3 domain-binding glutamic acid-rich-like protein 3
- SLC16A1 (1p13)
- SPSB1: SPRY domain-containing SOCS box protein 1
- STIL (1p33)
- SYCP1: Synaptonemal complex protein 1
- SZT2: Seizure threshold 2 homolog
- TACSTD2: tumor-associated calcium signal transducer 2
- TAL1 (1p33)
- TCEB3: Transcription elongation factor B polypeptide 3
- TGFBR3 (1p22)
- THRAP3 (1p34)
- TIE1 (1p34)
- TMCO4: encoding protein transmembrane and coiled-coil domains 4
- TMEM48: encoding protein nucleoporin NDC1
- TMEM50A: Transmembrane protein 50A
- TMEM59: Transmembrane protein 59
- TMEM69: Transmembrane protein 69
- TMEM201 encoding protein Transmembrane protein 201
- TMEM222: Transmembrane protein 222
- TOE1: Target of EGR1 protein 1
- TRAPPC3: Trafficking protein particle complex subunit 3
- TRIT1: tRNA isopentenyltransferase, mitochondrial
- TSHB: thyroid stimulating hormone, beta
- TTC39A: Tetratricopeptide repeat 39A
- UBR4: E3 ubiquitin-protein ligase component n-recognin 4
- UROD: uroporphyrinogen decarboxylase (the gene for porphyria cutanea tarda)
- USP1 (1p31)
- USP48: Ubiquitin carboxyl-terminal hydrolase 48
- VAV3 (1p13)
- VPS13D: Vacuolar protein sorting-associated protein 13D
- VTCN1 (1p13)
- WARS2: Tryptophanyl-tRNA synthetase, mitochondrial
- WDR77 (1p13)
- YBX1 (1p34)
- ZCCHC17: zinc finger CCHC-type containing 17
- ZMYM1 encoding protein Zinc finger MYM-type containing 1
- ZNF436: Zinc finger protein 436
- ZYG11B encoding protein Zyg-11 family member B, cell cycle regulator
- ZZZ3: ZZ-type zinc finger-containing protein 3
q-arm[edit]
Partial list of the genes located on q-arm (long arm) of human chromosome 1:
- ABL2 (1q25)
- ADIPOR1 (1q32)
- AHCTF1: encoding protein ELYS
- AKT3 (1q43-44)
- ANGPTL1: Angiopoietin-related protein 1
- ARHGEF2 (1q22)
- ARID4B: encoding protein AT-rich interactive domain-containing protein 4B
- ARV1 encoding protein ARV1 homolog (S. cerevisiae)
- ARNT (1q21)
- ASPM (1q31): a brain size determinant
- ATF3 (1q32)
- ATP2B4 (1q32)
- BCL9 (1q21)
- C1orf21: chromosome 1 open reading frame 21
- MMTAP2 encoding protein Multiple myeloma tumor-associated protein 2
- TEX35: TEX35
- C1orf74: chromosome 1 open reading frame 74
- C2CD4D: C2 calcium-dependent domain-containing 4D
- CD5L: CD5 molecule like
- CENPL: Centromere protein L
- CENPF (1q41)
- CHTOP: Chromatin target of prmt1
- CNIH4: cornichon homolog 4
- CNST: Consortin
- CREG1: Cellular repressor of E1A stimulated genes 1
- CRP: C-reactive protein
- CRTC2 (1q21)
- CSRP1: Cysteine and glycine rich protein 1
- DDX59: DEAD-box helicase 59
- DPT: Dermatopontin
- DISC2, long non-coding RNA
- DUSP10 (1q41)
- DNAH14 encoding protein Dynein, axonemal, heavy chain 14
- ECM1 (1q21)
- EDEM3: ER degradation enhancing alpha-mannosidase like protein 3
- EGLN1 (1q42)
- ENAH (1q42)
- ESRRG (1q41)
- FAM20B: FAM20B, glycosaminoglycan xylosylkinase
- FAM63A: Family with sequence similarity 63, member A
- FAM78B: family with sequence similarity 78, member B
- FAM129A: family with sequence similarity 129, member A
- FBXO28: F-box protein 28
- FCMR: Fc fragment of IgM receptor
- FCGR2B (1q23)
- FH (1q43)
- FMO3: flavin containing monooxygenase 3
- FRA1J encoding protein Fragile site, 5-azacytidine type, common, fra(1)(q12)
- GAS5 (1q25)
- GBA: glucosidase, beta; acid (includes glucosylceramidase) (gene for Gaucher disease)
- GBAP1: glucosylceramidase beta pseudogene 1
- GLC1A: gene for glaucoma
- GON4L: gon-4 like
- GPA33 (1q24)
- GPR37L1 G protein-coupled receptor 37 like 1
- HEATR1: HEAT repeat-containing protein 1
- HFE2: hemochromatosis type 2 (juvenile)
- HIST2H2AB: Histone 2A type 2-B
- HIST2H2BF: Histone H2B type 2-F
- HIST2H3PS2: Histone cluster 2, H3, pseudogene 2
- HIST3H2A: Histone H2A type 3
- HIST3H2BB: Histone H2B type 3-B
- HPC1: gene for prostate cancer
- HRM2: Hair, curly
- IGSF8 (1q23)
- INAVA: Innate immunity activator protein
- INTS3: Integrator complex subunit 3
- IRF6: gene for connective tissue formation
- KCNH1 (1q32)
- KIF14 (1q32)
- LEFTY1: Left-right determination factor 1
- LHX9 encoding protein LIM homeobox 9
- LMNA: lamin A/C
- LOC645166 encoding protein Lymphocyte-specific protein 1 pseudogene
- LYPLAL1: Lysophospholipase-like 1
- MAPKAPK2 (1q32)
- MIR194-1: microRNA 194-1
- MIR5008: microRNA 5008
- MPC2: Mitochondrial pyruvate carrier 2
- MOSC1: MOCO sulphurase C-terminal domain containing 1
- MOSC2: MOSC domain-containing protein 2, mitochondrial
- MPZ: myelin protein zero (Charcot–Marie–Tooth neuropathy 1B)
- MSTO1: misato 1
- MTR: 5-methyltetrahydrofolate-homocysteine methyltransferase
- NAV1: Neuron navigator 1
- NBPF16: Neuroblastoma breakpoint family, member 16
- NOC2L: Nucleolar complex protein 2 homolog
- NUCKS1: Nuclear ubiquitous casein and cyclin-dependent kinases substrate
- NVL: Nuclear valosin-containing protein-like
- OLFML2B: Olfactomedin-like 2B
- OPTC: Opticin
- OTUD7B: OTU domain-containing protein 7B
- PACERR encoding protein PTGS2 antisense NFKB1 complex-mediated expression regulator RNA
- PBX1 (1q23)
- PEA15 (1q23)
- PGDB5: PiggyBac transposable element derived 5
- PIAS3 (1q21)
- PI4KB: Phosphatidylinositol 4-kinase beta
- PIP5K1A (1q21): Phosphatidylinositol-4-phosphate 5-kinase type-1 alpha
- PLA2G4A (1q31)
- PPOX: protoporphyrinogen oxidase
- PRCC (1q23)
- PRR9 encoding protein Proline rich 9
- PSEN2 (1q42): presenilin 2 (Alzheimer disease 4)
- PTGS2 (1q31)
- PTPN14 (1q32-41)
- PTPN7 (1q32)
- RABIF: RAB interacting factor
- RASSF5 (1q32)
- RGS2 (1q31)
- RN5S1@: RNA, 5S ribosomal 1q42 cluster
- RPS27 (1q21)
- SCAMP3: Secretory carrier-associated membrane protein 3
- SDHC (1q23)
- SELE (1q24)
- SHC1 (1q21)
- SLC39A1 (1q21)
- SLC50A1: Solute carrier family 50 member 1
- SMCP: Sperm mitochondrial-associated cysteine-rich protein
- SMG7: nonsense mediated mRNA decay factor
- SMYD3 (1q44)
- SPG23
- SPRR1A: Cornifin-A
- SPRR1B: Cornifin-B
- SPRR2A: Small proline rich protein 2A
- SPRTN: Spartan
- TARBP1: TAR (HIV-1) RNA-binding protein 1
- TBCE: Tubulin-specific chaperone E
- THBS3: Thrombospondin 3
- TMCO1: Transmembrane and coiled-coil domain-containing protein 1
- TMEM9: Transmembrane protein 9
- TMEM63A: Transmembrane protein 63A
- TMEM81: Transmembrane protein 81
- TNFSF18 (1q25)
- TNN (1q25)
- TNNT2: cardiac troponin T2
- TOR1AIP1: Torsin-1A-interacting protein 1
- TP53BP2 (1q41)
- TRE-CTC1-5: Transfer RNA-Glu (CTC) 1-5
- TRP (1q31)
- UAP1: UDP-N-acetylhexosamine pyrophosphorylase
- USH2A: Usher syndrome 2A (autosomal recessive, mild)
- USF1 (1q23)
- VPS45: Vacuolar protein sorting-associated protein 45
- VPS72: Vacuolar protein sorting-associated protein 72
- YY1AP1: YY1-associated protein 1
- ZBED6: zinc finger, BED-type containing 6
- ZC3H11A: Zinc finger CCCH domain-containing protein 11A
- ZNF687: Zinc finger protein 687
- ZNF648 encoding protein Zinc finger protein 648
- ZNF692: Zinc finger protein 692
- ZNF695: Zinc finger protein 695
Diseases and disorders[edit]
There are 890 known diseases related to this chromosome.[citation needed] Some of these diseases are hearing loss, Alzheimer's disease, glaucomaand breast cancer. Rearrangements and mutations of chromosome 1 are prevalent in cancer and many other diseases. Patterns of sequence variation reveal signals of recent selection in specific genes that may contribute to human fitness, and also in regions where no function is evident.
Complete monosomy (only having one copy of the entire chromosome) is invariably lethal before birth.[14] Complete trisomy (having three copies of the entire chromosome) is lethal within days after conception.[14] Some partial deletions and partial duplications produce birth defects.
The following diseases are some of those related to genes on chromosome 1 (which contains the most known genetic diseases of any human chromosome):
- 1q21.1 deletion syndrome
- 1q21.1 duplication syndrome
- Alzheimer's disease
- Breast cancer
- Brooke Greenberg Disease (Syndrome X)
- Carnitine palmitoyltransferase II deficiency
- Charcot–Marie–Tooth disease, types 1 and 2
- collagenopathy, types II and XI
- congenital hypothyroidism
- Ehlers-Danlos syndrome
- Factor V Leiden thrombophilia
- Familial adenomatous polyposis
- galactosemia
- Gaucher disease
- Gaucher-like disease
- Gelatinous drop-like corneal dystrophy
- Glaucoma
- Hearing loss, autosomal recessive deafness 36
- Hemochromatosis
- Hepatoerythropoietic porphyria
- Homocystinuria
- Hutchinson Gilford progeria syndrome
- 3-hydroxy-3-methylglutaryl-CoA lyase deficiency
- Hypertrophic cardiomyopathy, autosomal dominant mutations of TNNT2; hypertrophy usually mild; restrictive phenotype may be present; may carry high risk of sudden cardiac death
- maple syrup urine disease
- medium-chain acyl-coenzyme A dehydrogenase deficiency
- Microcephaly
- Muckle-Wells Syndrome
- Nonsyndromic deafness
- Oligodendroglioma
- Parkinson disease
- Pheochromocytoma
- porphyria
- porphyria cutanea tarda
- popliteal pterygium syndrome
- prostate cancer
- Stickler syndrome
- TAR syndrome
- trimethylaminuria
- Usher syndrome
- Usher syndrome type II
- Van der Woude syndrome
- Variegate porphyria
 Protoporphyrinogen IXSpecialtyEndocrinology Symptomsskin problems, Enzyme difficiency.Causesgenetic mutations.Treatmentliver transplants.
Protoporphyrinogen IXSpecialtyEndocrinology Symptomsskin problems, Enzyme difficiency.Causesgenetic mutations.Treatmentliver transplants.Erythropoietic protoporphyria (or commonly called EPP) is a form of porphyria, which varies in severity and can be very painful. It arises from a deficiency in the enzyme ferrochelatase, leading to abnormally high levels of protoporphyrin in the red blood cells (erythrocytes), plasma, skin, and liver.[2] The severity varies significantly from individual to individual.
A clinically similar form of porphyria, known as X-Linked dominant protoporphyria, was identified in 2008.[3]
Haematin (also known as hematin, ferriheme, hematosin, hydroxyhemin, oxyheme, phenodin, or oxyhemochromogen) is a dark bluish or brownish pigment containing iron in the ferric state, obtained by the oxidation of haem.[1]
Haematin inhibits the synthesis of porphyrin, and stimulates the synthesis of globin. It is a component of cytochromes and peroxidases, and is also used as a reagent.[2]
https://en.wikipedia.org/wiki/Haematin
The globins are a superfamily of heme-containing globular proteins, involved in binding and/or transporting oxygen. These proteins all incorporate the globin fold, a series of eight alpha helical segments. Two prominent members include myoglobin and hemoglobin. Both of these proteins reversibly bind oxygen via a heme prosthetic group. They are widely distributed in many organisms.[2]
https://en.wikipedia.org/wiki/Globin
https://en.wikipedia.org/wiki/Globulin
https://en.wikipedia.org/wiki/Globular_protein
Crohn's resistance was a result of the absence of a receptor, which prevent the HIV from infecting CD4 present on the exterior of the white blood cells. The absence of such receptors, or rather the shortening of them to the point of being inoperable, is known as the delta 32 mutation.[4] This mutation is linked to groups of people that have been exposed to HIV but remain uninfected such as some offspring of HIV positive mothers, health officials, and sex workers.[5]
C-C chemokine receptor type 5, also known as CCR5 or CD195, is a protein on the surface of white blood cells that is involved in the immune system as it acts as a receptor for chemokines. This is the process by which T cells are attracted to specific tissue and organ targets. Many strains of HIV use CCR5 as a co-receptor to enter and infect host cells. A few individuals carry a mutation known as CCR5-Δ32 in the CCR5 gene, protecting them against these strains of HIV.[citation needed]
In humans, the CCR5 gene that encodes the CCR5 protein is located on the short (p) arm at position 21 on chromosome 3. A cohort study, from June 1981 to October 2016, looked into the correlation between the delta 32 deletion and HIV resistance, and found that homozygous carriers of the delta 32 mutation are resistant to M-tropic strains of HIV-1 infection.[9] Certain populations have inherited the Delta 32 mutation resulting in the genetic deletion of a portion of the CCR5 gene.[10][excessive citations]
In 2019, it was discovered that the mutation of TNPO3 that is the cause of type 1F limb-girdle muscular dystrophy (LGMD1F), also causes innate resistance to HIV-1.[11] The gene TNP03 was known to be involved into virus transportation into the infected cells. Blood samples from a family affected by LGMD1F showed a resistance to HIV infection. While the CCR5Δ32 deletion blocks the entry of virus strains that use the CCR5 receptor, the TNPO3 mutation causing LGMD1F blocks the CXCR4 receptor, making it effective on different HIV-1 strains, due to HIV tropism.[citation needed]
Cytotoxic T-lymphocytes (CTLs) provide a protective reaction against HIV when consistent exposure to the virus is present. Sex workers are found to have these CTLs within genital mucus, preventing the spread of HIV within heterosexual transmission. While creating a protective seal, CTLs become ineffective when lapses in HIV exposure occur, which leads to the possibility of CTLs only being an indicator of other genetic resistances towards HIV, such as immunoglobulin A responses within vaginal fluids.[5][12]
Chimpanzees in African countries have been found to develop AIDS at a slower rate than humans. This resistance is not due to the primate's ability to control the virus in a manner that is substantially more effective than humans, but rather because of the lack of tissues created within the body that typically progress HIV to AIDS. The chimpanzees also lack CD4 T cells and immune activation that is required for the spread of HIV.[12]
Another method used by the Kiem lab was the release of zinc finger nuclease (ZFN), which identifies specific sections of DNA to cause a break in the double helix. These ZFNs were used to target CCR5 in order to delete the protein, halting the course of the infection.[13]
While the delta mutation has been observed to prevent HIV in specific populations, it has shown little to no effect between healthy individuals and those who are infected with HIV among Iranian populations. This is attributed to individuals being heterozygous for the mutation, which prevents the delta mutation from effectively prohibiting HIV from entering immune cells.[15]
https://en.wikipedia.org/wiki/Innate_resistance_to_HIV
With in vitro transcription techniques three suggested ORFs on chromosome 3 (gag), 6 (pro) and 7 (env) were detected and further analyzed revealing that the ORF on chromosome 7q21.2 uniquely encoded a glycosylated Env protein.[12] Performing RealTime RT-PCR on adrenal gland, bone marrow, cerebellum, whole brain, fetal brain, fetal liver, heart, kidney, liver, lung, placenta, prostate, salivary gland, skeletal muscle, spinal cord, testis, thymus, thyroid gland, trachea, and uterus cells revealed 22 complete HERV-W families on chromosomes 1–3, 5–8, 10–12, 15, 19 and X.[6]
In silico expression data revealed that these HERV-W elements are randomly expressed in various tissues (brain, mammary gland, cerebrum, skin, testis, eye, embroyonic tissue, pancreatic islet, pineal gland, endocrine, retina, adipose tissue, placenta and muscle).[6]
Further, human tissues that lack some sort of HERV expression could not be found which suggests that HERVs are permanent members of the human transcriptome.[13] Although expression of HERV-W is prevalent in the whole body there are two tissues whose expression levels are higher than the rest. The HERV-W derived element of chromosome 12p11.21 and 7q21.2 had 42 hits from the env gene in pancreatic islet tissues and 224 hits (11 gag, 41 pol, 164 env) in placenta, testis, and embryotic tissues, respectively. The HERV-W element on 7q21.2 encodes for ERVWE-1, which was named synctin-1.[14]
As a control a gene known to be hyperfusogenic, A-Rless, was transfected into the cell-line . Upon transfection of cells with this vector there was only a 6% fusion of cells as opposed to a 48% fusion with the HERV-W vector, thus revealing the gene encoded by HERV-W env is a highly fusogenic membrane glycoprotein.[15]
Furthermore, HERV-W env glycoprotein has been shown to contain an immunosuppressive region.[21] This immunosuppressive nature of synctin-1 and/or synctin-2 (HERV-W) may be key in creating an immunologic barrier between the mother and the fetus.[22] Since the fetus only share half of the mother's DNA it is critical that the mother's immune system does not reject or attack the fetus.[23]
https://en.wikipedia.org/wiki/Human_Endogenous_Retrovirus-W
Sharon Petersins Family Geneti
https://en.wikipedia.org/wiki/Catarrhini
However, using a luciferase reporter gene assay HERV-Ws that have incomplete LTR's were still found to have promoter activity. This suggests that the transcription of HERV's can be activated not just by LTR- directed transcription but also by transcriptional leakage.[25] Meaning if a nearby gene is being transcribed the transcription factors and polymerase can just keep moving along the DNA reaching the nearby HERV, where they can then transcribe it. In fact by doing a Chip-seq analysis of HERV-W LTR's it was found that ¼ of HERV-W LTR's can be bound by transcription factor p56 (ENCODE Project). This indicates a reason behind HERV-W's cell specific expression.
https://en.wikipedia.org/wiki/Human_Endogenous_Retrovirus-W
| Chromosome | Length (mm) | Base pairs | Variations | Protein- coding genes | Pseudo- genes | Total long ncRNA | Total small ncRNA | miRNA | rRNA | snRNA | snoRNA | Misc ncRNA | Links | Centromere position (Mbp) | Cumulative (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 85 | 248,956,422 | 12,151,146 | 2058 | 1220 | 1200 | 496 | 134 | 66 | 221 | 145 | 192 | EBI | 125 | 7.9 |
| 2 | 83 | 242,193,529 | 12,945,965 | 1309 | 1023 | 1037 | 375 | 115 | 40 | 161 | 117 | 176 | EBI | 93.3 | 16.2 |
| 3 | 67 | 198,295,559 | 10,638,715 | 1078 | 763 | 711 | 298 | 99 | 29 | 138 | 87 | 134 | EBI | 91 | 23 |
| 4 | 65 | 190,214,555 | 10,165,685 | 752 | 727 | 657 | 228 | 92 | 24 | 120 | 56 | 104 | EBI | 50.4 | 29.6 |
| 5 | 62 | 181,538,259 | 9,519,995 | 876 | 721 | 844 | 235 | 83 | 25 | 106 | 61 | 119 | EBI | 48.4 | 35.8 |
| 6 | 58 | 170,805,979 | 9,130,476 | 1048 | 801 | 639 | 234 | 81 | 26 | 111 | 73 | 105 | EBI | 61 | 41.6 |
| 7 | 54 | 159,345,973 | 8,613,298 | 989 | 885 | 605 | 208 | 90 | 24 | 90 | 76 | 143 | EBI | 59.9 | 47.1 |
| 8 | 50 | 145,138,636 | 8,221,520 | 677 | 613 | 735 | 214 | 80 | 28 | 86 | 52 | 82 | EBI | 45.6 | 52 |
| 9 | 48 | 138,394,717 | 6,590,811 | 786 | 661 | 491 | 190 | 69 | 19 | 66 | 51 | 96 | EBI | 49 | 56.3 |
| 10 | 46 | 133,797,422 | 7,223,944 | 733 | 568 | 579 | 204 | 64 | 32 | 87 | 56 | 89 | EBI | 40.2 | 60.9 |
| 11 | 46 | 135,086,622 | 7,535,370 | 1298 | 821 | 710 | 233 | 63 | 24 | 74 | 76 | 97 | EBI | 53.7 | 65.4 |
| 12 | 45 | 133,275,309 | 7,228,129 | 1034 | 617 | 848 | 227 | 72 | 27 | 106 | 62 | 115 | EBI | 35.8 | 70 |
| 13 | 39 | 114,364,328 | 5,082,574 | 327 | 372 | 397 | 104 | 42 | 16 | 45 | 34 | 75 | EBI | 17.9 | 73.4 |
| 14 | 36 | 107,043,718 | 4,865,950 | 830 | 523 | 533 | 239 | 92 | 10 | 65 | 97 | 79 | EBI | 17.6 | 76.4 |
| 15 | 35 | 101,991,189 | 4,515,076 | 613 | 510 | 639 | 250 | 78 | 13 | 63 | 136 | 93 | EBI | 19 | 79.3 |
| 16 | 31 | 90,338,345 | 5,101,702 | 873 | 465 | 799 | 187 | 52 | 32 | 53 | 58 | 51 | EBI | 36.6 | 82 |
| 17 | 28 | 83,257,441 | 4,614,972 | 1197 | 531 | 834 | 235 | 61 | 15 | 80 | 71 | 99 | EBI | 24 | 84.8 |
| 18 | 27 | 80,373,285 | 4,035,966 | 270 | 247 | 453 | 109 | 32 | 13 | 51 | 36 | 41 | EBI | 17.2 | 87.4 |
| 19 | 20 | 58,617,616 | 3,858,269 | 1472 | 512 | 628 | 179 | 110 | 13 | 29 | 31 | 61 | EBI | 26.5 | 89.3 |
| 20 | 21 | 64,444,167 | 3,439,621 | 544 | 249 | 384 | 131 | 57 | 15 | 46 | 37 | 68 | EBI | 27.5 | 91.4 |
| 21 | 16 | 46,709,983 | 2,049,697 | 234 | 185 | 305 | 71 | 16 | 5 | 21 | 19 | 24 | EBI | 13.2 | 92.6 |
| 22 | 17 | 50,818,468 | 2,135,311 | 488 | 324 | 357 | 78 | 31 | 5 | 23 | 23 | 62 | EBI | 14.7 | 93.8 |
| X | 53 | 156,040,895 | 5,753,881 | 842 | 874 | 271 | 258 | 128 | 22 | 85 | 64 | 100 | EBI | 60.6 | 99.1 |
| Y | 20 | 57,227,415 | 211,643 | 71 | 388 | 71 | 30 | 15 | 7 | 17 | 3 | 8 | EBI | 10.4 | 100 |
| mtDNA | 0.0054 | 16,569 | 929 | 13 | 0 | 0 | 24 | 0 | 2 | 0 | 0 | 0 | EBI | N/A | 100 |
| total | 3,088,286,401 | 155,630,645 | 20412 | 14600 | 14727 | 5037 | 1756 | 532 | 1944 | 1521 | 2213 |
Original analysis published in the Ensembl database at the European Bioinformatics Institute (EBI) and Wellcome Trust Sanger Institute. Chromosome lengths estimated by multiplying the number of base pairs by 0.34 nanometers (distance between base pairs in the most common structure of the DNA double helix; a recent estimate of human chromosome lengths based on updated data reports 205.00 cm for the diploid male genome and 208.23 cm for female, corresponding to weights of 6.41 and 6.51 picograms (pg), respectively[27]). Number of proteins is based on the number of initial precursor mRNA transcripts, and does not include products of alternative pre-mRNA splicing, or modifications to protein structure that occur after translation.
Variations are unique DNA sequence differences that have been identified in the individual human genome sequences analyzed by Ensembl as of December 2016. The number of identified variations is expected to increase as further personal genomes are sequenced and analyzed. In addition to the gene content shown in this table, a large number of non-expressed functional sequences have been identified throughout the human genome (see below). Links open windows to the reference chromosome sequences in the EBI genome browser.
Small non-coding RNAs are RNAs of as many as 200 bases that do not have protein-coding potential. These include: microRNAs, or miRNAs (post-transcriptional regulators of gene expression), small nuclear RNAs, or snRNAs (the RNA components of spliceosomes), and small nucleolar RNAs, or snoRNA (involved in guiding chemical modifications to other RNA molecules). Long non-coding RNAs are RNA molecules longer than 200 bases that do not have protein-coding potential. These include: ribosomal RNAs, or rRNAs (the RNA components of ribosomes), and a variety of other long RNAs that are involved in regulation of gene expression, epigenetic modifications of DNA nucleotides and histone proteins, and regulation of the activity of protein-coding genes. Small discrepancies between total-small-ncRNA numbers and the numbers of specific types of small ncNRAs result from the former values being sourced from Ensembl release 87 and the latter from Ensembl release 68.
The number of genes in the human genome is not entirely clear because the function of numerous transcripts remains unclear. This is especially true for non-coding RNA. The number of protein-coding genes is better known but there are still on the order of 1,400 questionable genes which may or may not encode functional proteins, usually encoded by short open reading frames.
| Gencode[29] | Ensembl[30] | Refseq[31] | CHESS[32] | |
|---|---|---|---|---|
| protein-coding genes | 19,901 | 20,376 | 20,345 | 21,306 |
| lncRNA genes | 15,779 | 14,720 | 17,712 | 18,484 |
| antisense RNA | 5501 | 28 | 2694 | |
| miscellaneous RNA | 2213 | 2222 | 13,899 | 4347 |
| Pseudogenes | 14,723 | 1740 | 15,952 | |
| total transcripts | 203,835 | 203,903 | 154,484 | 328,827 |
https://en.wikipedia.org/wiki/Human_genome
| Tay-Sachs | 1:3600 births in Ashkenazi Jews | HEXA gene (on chromosome 15) Group Var |
| Mucolipidosis type IV | 1:90 to 1:100 in Ashkenazi Jews | MCOLN1 PetersenSmithUSAMisterGenModCloHybrJJ |
| Other conditions | ||
|---|---|---|
| Cystic fibrosis | 1:2500 | CFTR |
| Duchenne muscular dystrophy | 1:3500 boys | Dystrophin |
| Becker muscular dystrophy | 1.5-6:100000 males | DMD |
| Beta thalassemia | 1:100000 | HBB |
| Congenital adrenal hyperplasia | 1:280 in Native Americans and Yupik Eskimos 1:15000 in American Caucasians | CYP21A2 |
| Glycogen storage disease type I | 1:100000 births in America | G6PC |
| Maple syrup urine disease | 1:180000 in the U.S. 1:176 in Mennonite/Amish communities 1:250000 in Austria | BCKDHA, BCKDHB, DBT, DLD |
| Niemann–Pick disease, SMPD1-associated | 1,200 cases worldwide | SMPD1 |
| Usher syndrome | 1:23000 in the U.S. 1:28000 in Norway 1:12500 in Germany | CDH23, CLRN1, DFNB31, GPR98, MYO7A, PCDH15, USH1C, USH1G, USH2A |
| Disorder | Prevalence | Chromosome or gene involved |
|---|---|---|
| Chromosomal conditions | ||
| Down syndrome | 1:600 | Chromosome 21 |
| Klinefelter syndrome | 1:500–1000 males | Additional X chromosome |
| Turner syndrome | 1:2000 females | Loss of X chromosome |
| Sickle cell anemia | 1 in 50 births in parts of Africa; rarer elsewhere | β-globin (on chromosome 11) |
| Bloom syndrome | 1:48000 Ashkenazi Jews | BLM |
| Cancers | ||
| Breast/Ovarian cancer(susceptibility) | ~5% of cases of these cancer types | BRCA1, BRCA2 |
| FAP (hereditary nonpolyposis coli) | 1:3500 | APC |
| Lynch syndrome | 5–10% of all cases of bowel cancer | MLH1, MSH2, MSH6, PMS2 |
| Fanconi anemia | 1:130000 births | FANCC |
| Neurological conditions | ||
| Huntington disease | 1:20000 | Huntingtin |
| Alzheimer disease ‐ early onset | 1:2500 | PS1, PS2, APP |
| Tay-Sachs | 1:3600 births in Ashkenazi Jews | HEXA gene (on chromosome 15) |
| Canavan disease | 2.5% Eastern European Jewish ancestry | ASPA gene (on chromosome 17) |
| Familial dysautonomia | 600 known cases worldwide since discovery | IKBKAP gene (on chromosome 9) |
| Fragile X syndrome | 1.4:10000 in males, 0.9:10000 in females | FMR1 gene (on X chromosome) |
| Mucolipidosis type IV | 1:90 to 1:100 in Ashkenazi Jews | MCOLN1 |




No comments:
Post a Comment