Rhinitis, also known as coryza,[3] is irritation and inflammation of the mucous membrane inside the nose. Common symptoms are a stuffy nose, runny nose, sneezing, and post-nasal drip.[4]
The inflammation is caused by viruses, bacteria, irritants or allergens. The most common kind of rhinitis is allergic rhinitis, [5] which is usually triggered by airborne allergens such as pollen and dander.[6] Allergic rhinitis may cause additional symptoms, such as sneezing and nasal itching, coughing, headache,[7] fatigue, malaise, and cognitive impairment.[8][9] The allergens may also affect the eyes, causing watery, reddened, or itchy eyes and puffiness around the eyes.[7] The inflammation results in the generation of large amounts of mucus, commonly producing a runny nose, as well as a stuffy nose and post-nasal drip. In the case of allergic rhinitis, the inflammation is caused by the degranulation of mast cells in the nose. When mast cells degranulate, they release histamine and other chemicals,[10] starting an inflammatory process that can cause symptoms outside the nose, such as fatigue and malaise.[11] In the case of infectious rhinitis, it may occasionally lead to pneumonia, either viral or bacterial. Sneezing also occurs in infectious rhinitis to expel bacteria and viruses from the respiratory tract.
Rhinitis is very common. Allergic rhinitis is more common in some countries than others; in the United States, about 10–30% of adults are affected annually.[12] Mixed rhinitis (MR) refers to patients with nonallergic rhinitis and allergic rhinitis. MR is a specific rhinitis subtype. It may represent between 50 and 70% of all AR patients. However, true prevalence of MR has not been confirmed yet.[13]
https://en.wikipedia.org/wiki/Rhinitis
Leucosis is a leukemia-like malignant viral disease that is found in animals, particularly poultry and cattle.
Types of leucosis[edit]
- Bovine leucosis
- Enzootic bovine leucosis, caused by bovine leukemia virus.[1]
- Sporadic bovine leucosis
- Calf lymphosarcoma
- Leucosis in pig
- Leucosis in horses
- Leucosis in sheep [2]
- Feline leucosis
- Avian leucosis and related diseases
- Avian sarcoma leukosis virus
- Lymphoid leucosis
- Erythroblastosis
- Osteopetrosis
- Myeloblastose
- Myelocytomatosis[3]
FeLV was first described in cats in 1964.[25] The disease was originally associated with leukemia; however, it was later realized that the initial signs are generally anemia and immunosuppression.[25] The first diagnostic test became available in 1973, which led to a "test and elimination" regime, dramatically reducing the number of infected cats in the general population.[26] The first vaccine became available in 1986.[26]
Progression
[edit]
The disease has a wide range of effects. The cat can fight off the infection and become totally immune, can become a healthy carrier that never gets sick itself but can infect other cats, or a mid-level case in which the cat has a compromised immune system.[citation needed] Nevertheless, the development of lymphomas is considered the final stage of the disease. Although it is thought that virus protein has to be present to induce lymphomas in cats, newer evidence shows that a high percentage of FeLV-Antigen negative lymphomas contain FeLV-DNA, indicating a "hit-and-run" mechanism of virus-induced tumor development.[5]
Once the virus has entered the cat, there are six stages to a FeLV infection[citation needed]:
- Stage One: The virus enters the cat, usually through the pharynx where it infects the epithelial cells and infects the tonsilar B-lymphocytes and macrophages. These white blood cells then filter down to the lymph nodes and begin to replicate.
- Stage Two: The virus enters the blood stream and begins to distribute throughout the body.
- Stage Three: The lymphoid system (which produces antibodies to attack infected and cancerous cells) becomes infected, with further distribution throughout the body.
- Stage Four: The main point in the infection- where the virus can take over the body's immune system and cause viremia. During this stage the hemolymphatic system and intestines become infected.
If the cat's immune system does not fight off the virus, then it progresses to:
- Stage Five: The bone marrow becomes infected. At this point, the virus will stay with the cat for the rest of its life. In this phase, the virus replicates and is released four to seven days later in infected neutrophils, and sometimes lymphocytes, monocytes, and eosinophils (all white blood cells formed in the bone marrow).
- Stage Six: The cat's body is overwhelmed by infection and mucosal and glandular epithelial cells (tissue that forms a thin protective layer on exposed bodily surfaces and forms the lining of internal cavities, ducts, and organs) become infected. The virus replicates in epithelial tissues including salivary glands, oropharynx, stomach, esophagus, intestines, trachea, nasopharynx, renal tubules, bladder, pancreas, alveolar ducts, and sebaceous ducts from the muzzle.
Comparison with feline immunodeficiency virus
[edit]
FeLV and feline immunodeficiency virus (FIV) are sometimes mistaken for one another, though the viruses differ in many ways. Although they are both in the same retroviral subfamily (Orthoretrovirinae), they are classified in different genera (FeLV is a gamma-retrovirus and FIV is a lentivirus like HIV-1). Their shapes are quite different: FeLV is more circular while FIV is elongated. The two viruses are also quite different genetically, and their protein coats differ in size and composition. Although many of the diseases caused by FeLV and FIV are similar, the specific ways in which they are caused also differ. Also, while the feline leukemia virus may cause symptomatic illness in an infected cat, an FIV infected cat can remain completely asymptomatic its entire lifetime.[citation needed]
See also[edit]
https://en.wikipedia.org/wiki/Feline_leukemia_virus
Endogenous Retrovirus
See also[edit]
icon Viruses portal
Avian sarcoma leukosis virus (ASLV)
Endogenous viral element
ERV3
HERV-FRD
Horizontal gene transfer
Jaagsiekte sheep retrovirus (JSRV)
Koala retrovirus (KoRV)
Mouse mammary tumor virus (MMTV)
Murine leukemia virus (MLV) and xenotropic murine leukemia virus-related virus (XMRV)
Paleovirology
In mammals, intact env proteins called syncytins are responsible for the formation and function of syncytiotrophoblasts.[15] These multinucleated cells are mainly responsible for maintaining nutrient exchange and separating the fetus from the mother's immune system.[15] It has been suggested that the selection and fixation of these proteins for this function have played a critical role in the evolution of viviparity.[30]
https://en.wikipedia.org/wiki/Endogenous_retrovirus
https://en.wikipedia.org/wiki/Pospiviroidae
https://en.wikipedia.org/wiki/Avsunviroidae
https://en.wikipedia.org/wiki/Defective_interfering_particle
https://en.wikipedia.org/wiki/Fungal_prion
https://en.wikipedia.org/wiki/Mobilome
Self Replicating ORganic Structures; Genetic Transplantation Vectors at GMO genetically modified organism food crop etc.
https://en.wikipedia.org/wiki/Endogenous_retrovirus
https://en.wikipedia.org/wiki/Transpoviron
https://en.wikipedia.org/wiki/Extrachromosomal_DNA
Extrachromosomal circular DNA (eccDNA) are circular DNA found in human, plant and animal cells in addition to chromosomal DNA. eccDNA originate from chromosomal DNA and can be from 50 base pairs to several mega-base pairs in length and encode regulatory elements and several full genes.
eccDNA was first discovered in 1964 by Alix Bassel and Yasuo Hoota[1] in wheat nuclei and boar sperm.[2] Since then, eccDNA has been observed in almost all organisms from plants, yeast, C. elegans, frogs, mice, chicken, birds, and humans.[3][4][5][6][7][8][9] eccDNA molecules originate in normal cells and are a by product of programmed DNA recombination events; such as V(D)J recombination.[10][9] Moreover, eccDNA production seems to be cell-type specific in somatic cells.[9]
A subtype of eccDNA, such as ecDNA, ribosomal DNA locus (Extrachromosomal rDNA circle), and double minutes have been associated with genomic instability. Double minute ecDNAs are fragments of extrachromosomal DNA, which were originally observed in a large number of human tumors including breast, lung, ovary, colon, and most notably, neuroblastoma. They are a manifestation of gene amplification during the development of tumors, which give the cells selective advantages for growth and survival. Double minutes, like actual chromosomes, are composed of chromatinand replicate in the nucleus of the cell during cell division. Unlike typical chromosomes, they are composed of circular fragments of DNA, up to only a few million base pairs in size and contain no centromere or telomere.
See also[edit]
https://en.wikipedia.org/wiki/Extrachromosomal_circular_DNA#Role_of_ecDNA_in_cancer
Plasmodium is a genus of unicellular eukaryotes that are obligate parasites of vertebrates and insects. The life cycles of Plasmodium species involve development in a blood-feeding insect host which then injects parasites into a vertebrate host during a blood meal. Parasites grow within a vertebrate body tissue (often the liver) before entering the bloodstream to infect red blood cells. The ensuing destruction of host red blood cells can result in disease, called malaria. During this infection, some parasites are picked up by a blood-feeding insect (mosquitoes in majority cases), continuing the life cycle.[1]
https://en.wikipedia.org/wiki/Plasmodium
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently.
https://en.wikipedia.org/wiki/Plasmid
A phagemid or phasmid is a DNA-based cloning vector, which has both bacteriophage and plasmid properties.[1] These vectors carry, in addition to the origin of plasmid replication, an origin of replication derived from bacteriophage. Unlike commonly used plasmids, phagemid vectors differ by having the ability to be packaged into the capsid of a bacteriophage, due to their having a genetic sequence that signals for packaging. Phagemids are used in a variety of biotechnology applications; for example, they can be used in a molecular biology technique called "Phage Display".[2]
Filamentous phages retard bacterial growth but, contrasting with the lambda phage and the T7 phage, are not generally lytic. Helper phages are usually engineered to package less efficiently (via a defective phage origin of replication)[4] than the phagemid so that the resultant phage particles contain predominantly phagemid DNA. F1 Filamentous phage infection requires the presence of a pilus so only bacterial hosts containing the F-plasmid or its derivatives can be used to generate phage particles.
Prior to the development of cycle sequencing, phagemids were used to generate single stranded DNA template for sequencing purposes. Today phagemids are still useful for generating templates for site-directed mutagenesis. Detailed characterisation of the filamentous phage life cycle and structural features lead to the development of phage display technology, in which a range of peptides and proteins can be expressed as fusions to phage coat proteins and displayed on the viral surface. The displayed peptides and polypeptides are associated with the corresponding coding DNA within the phage particle and so this technique lends itself to the study of protein-protein interactions and other ligand/receptor combinations.
https://en.wikipedia.org/wiki/Phagemid
https://en.wikipedia.org/wiki/Phagemid
Dinodnavirus is a genus of viruses that infect dinoflagellates.[1] This genus belongs to the clade of nucleocytoplasmic large DNA viruses. The name is derived from 'dino' (dinoflagellate) and DNA (from its genome).
The only species in the genus is Heterocapsa circularisquama DNA virus 01.[2]
https://en.wikipedia.org/wiki/Dinodnavirus
Rhizidiovirus is a genus of viruses. Stramenopiles[1] (fungi and hyphochytridiomycota) serve as natural hosts. There is only one species in this genus: Rhizidiomyces virus.[2][3]
https://en.wikipedia.org/wiki/Rhizidiovirus
Plasmaviridae is a family of bacteria-infecting viruses. Acholeplasma species serve as natural hosts. There is one genus in the family, Plasmavirus, which contains one species: Acholeplasma virus L2.[1][2][3] All viruses known in this family have been isolated from species in the class Mollicutes.
This family is poorly studied and little is known about the diversity and biology of these viruses.
https://en.wikipedia.org/wiki/Plasmaviridae
A polydnavirus /pɒˈlɪdnəvaɪrəs/ (PDV) is a member of the family Polydnaviridae of insect viruses. There are two genera in the family: Bracovirus and Ichnovirus. Polydnaviruses form a symbiotic relationship with parasitoid wasps (ichnoviruses (IV) occur in ichneumonid wasps species and bracoviruses (BV) in braconid wasps), but these wasps are themselves parasitic on Lepidoptera (moths and butterflies).[1][2] Little or no sequence homology exists between BV and IV, suggesting that the two genera have been evolving independently for a long time.
https://en.wikipedia.org/wiki/Polydnavirus
Finnlakeviridae is a family of bacterial viruses that is not assigned to any higher taxonomic ranks. The family contains a single genus, Finnlakevirus, which contains a single species, Flavobacterium virus FLiP.[1][2] This virus was isolated in 2010, with its gram-negative host bacterium, from Lake Jyväsjärvi, a boreal freshwater habitat in Central Finland, and is the first described single-stranded DNA virus with an internal membrane.[2]
https://en.wikipedia.org/wiki/Finnlakeviridae
A nanobe[pronunciation?] is a tiny filamental structure first found in some rocks and sediments. Some scientists hypothesize that nanobes are the smallest form of life, 1/10 the size of the smallest known bacteria.[1]
No conclusive evidence exists that these structures are, or are not, living organisms, so their classification is controversial.
The 1996 discovery of nanobes was published in 1998[2] by Philippa Uwins et al.,[3] from the University of Queensland, Australia. They were found growing from rock samples (both full-diameter and sidewall cores) of Jurassic and Triassic sandstones, originally retrieved from an unspecified number of oil exploration wells off Australia's west coast. Depths of retrieval were between 3,400 metres (2.1 mi) and 5,100 metres (3.2 mi) below the sea bed. While Uwins et al. present assertions against it, they do not exclude the possibility that the nanobes are from a surface contaminant, not from the rock units cited.
The smallest are just 20 nanometers in diameter. Some researchers believe that these structures are crystal growths, but the staining of these structures with dyes that bind to DNA might indicate that they are living organisms.[4] They are similar to the structures found in ALH84001, a Mars meteorite found in the Antarctic. Nanobes are similar in size to nanobacteria, which are also structures that had been proposed to be extremely small living organisms. However, these two should not be confused. Nanobacteria were thought to be cellular organisms, while nanobes are hypothesized (by some) to be a previously unknown form of life or protocells.[citation needed]
https://en.wikipedia.org/wiki/Nanobe
Ultramicrobacteria are bacteria that are smaller than 0.1 μm3 under all growth conditions.[1][2][3] This term was coined in 1981, describing cocci in seawater that were less than 0.3 μm in diameter.[4] Ultramicrobacteria have also been recovered from soil and appear to be a mixture of Gram-positive, Gram-negative and cell-wall-lacking species.[5][2] Ultramicrobacteria possess a relatively high surface-area-to-volume ratio due to their small size, which aids in growth under oligotrophic (i.e. nutrient-poor) conditions.[2] The relatively small size of ultramicrobacteria also enables parasitism of larger organisms;[2] some ultramicrobacteria have been observed to be obligate or facultative parasites of various eukaryotes and prokaryotes.[1][2]One factor allowing ultramicrobacteria to achieve their small size seems to be genome minimization[1][2] such as in the case of the ultramicrobacterium P. ubique whose small 1.3 Mb genome is seemingly devoid of extraneous genetic elements like non-coding DNA, transposons, extrachromosomal elements etc.[2] However, genomic data from ultramicrobacteria is lacking[2] since the study of ultramicrobacteria, like many other prokaryotes, is hindered by difficulties in cultivating them.[3]
Ultramicrobacteria are commonly confused with ultramicrocells, the latter of which are the dormant, stress-resistant forms of larger cells that form under starvation conditions[1][2][7] (i.e. these larger cells downregulate their metabolism, stop growing and stabilize their DNA to create ultramicrocells that remain viable for years[1][8]) whereas the small size of ultramicrobacteria is not a starvation response and is consistent even under nutrient-rich conditions.[3]
The term "nanobacteria" is sometimes used synonymously with ultramicrobacteria in the scientific literature,[2] but ultramicrobacteria are distinct from the purported nanobacteria or "calcifying nanoparticles", which were proposed to be living organisms that were 0.1 μm in diameter.[9] These structures are now thought to be non-living,[10] and likely precipitated particles of inorganic material.[11][12]
See also[edit]
- L-form bacteria
- Mycoplasma – smallest known bacteria (300 nm)
- Nanoarchaeum – smallest known archaeum (400 nm)
- Nanobacteria – possible lifeforms smaller than bacteria (<200 nm)
- Nanobe – possible smallest lifeforms (20 nm)
- Pandoravirus – one of the largest known viruses (1000 nm)
- Parvovirus – smallest known viruses (18–28 nm)
- Pithovirus – largest known virus (1500 nm)
- Prion – smallest known infectious agent (≈10 nm)
- ND5 and MY14T – two aerobic, Gram-negative, rod-shaped bacteria[13]
https://en.wikipedia.org/wiki/Ultramicrobacteria
L-form bacteria, also known as L-phase bacteria, L-phase variants, and cell wall-deficient (CWD) bacteria, are strains of bacteria that lack cell walls.[1] They were first isolated in 1935 by Emmy Klieneberger-Nobel, who named them "L-forms" after the Lister Institute in London where she was working.[2]
Two types of L-forms are distinguished: unstable L-forms, spheroplasts that are capable of dividing, but can revert to the original morphology, and stable L-forms, L-forms that are unable to revert to the original bacteria.
Some parasitic species of bacteria, such as mycoplasma, also lack a cell wall,[3] but these are not considered L-forms since they are not derived from bacteria that normally have cell walls.[4]
https://en.wikipedia.org/wiki/L-form_bacteria
Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants and humans. As a member of the genusBacillus, B. subtilis is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. B. subtilis has historically been classified as an obligate aerobe, though evidence exists that it is a facultative anaerobe. B. subtilis is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies.[3]
| Bacillus subtilis | |
|---|---|
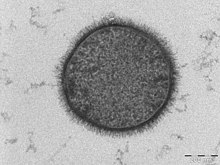 | |
| TEM micrograph of a B. subtilis cell in cross-section (scale bar = 200 nm) |
https://en.wikipedia.org/wiki/Bacillus_subtilis
Methotrexate (MTX), formerly known as amethopterin, is a chemotherapy agent and immune-system suppressant.[4] It is used to treat cancer, autoimmune diseases, and ectopic pregnancyand for medical abortions.[4] Types of cancers it is used for include breast cancer, leukemia, lung cancer, lymphoma, gestational trophoblastic disease, and osteosarcoma.[4] Types of autoimmune diseases it is used for include psoriasis, rheumatoid arthritis, and Crohn's disease.[4] It can be given by mouth or by injection.[4]
https://en.wikipedia.org/wiki/Methotrexate
L-forms can be generated in the laboratory from many bacterial species that usually have cell walls, such as Bacillus subtilis or Escherichia coli. This is done by inhibiting peptidoglycan synthesis with antibiotics or treating the cells with lysozyme, an enzyme that digests cell walls. The L-forms are generated in a culture medium that is the same osmolarity as the bacterial cytosol (an isotonic solution), which prevents cell lysis by osmotic shock.[2] L-form strains can be unstable, tending to revert to the normal form of the bacteria by regrowing a cell wall, but this can be prevented by long-term culture of the cells under the same conditions that were used to produce them – letting the wall-disabling mutations to accumulate by genetic drift.[6]
Some studies have identified mutations that occur, as these strains are derived from normal bacteria.[1][2]One such point mutation D92E is in an enzyme yqiD/ispA (P54383) involved in the mevalonate pathwayof lipid metabolism that increased the frequency of L-form formation 1,000-fold.[1] The reason for this effect is not known, but it is presumed that the increase is related to this enzyme's role in making a lipid important in peptidoglycan synthesis.
Another methodology of induction relies on nanotechnology and landscape ecology. Microfluidics devices can be built in order to challenge peptidoglycan synthesis by extreme spatial confinement. After biological dispersal through a constricted (sub-micrometre scale) biological corridor connecting adjacent micro habitat patches, L-form-like cells can be derived[7] using a microfluifics-based (synthetic) ecosystem implementing an adaptive landscape[8] selecting for shape-shifting phenotypes similar to L-forms.
https://en.wikipedia.org/wiki/L-form_bacteria
Significance and applications
[edit]
Some publications have suggested that L-form bacteria might cause diseases in humans,[9] and other animals[10] but, as the evidence that links these organisms to disease is fragmentary and frequently contradictory, this hypothesis remains controversial.[11][12] The two extreme viewpoints on this question are that L-form bacteria are either laboratory curiosities of no clinical significance or important but unappreciated causes of disease.[4]Research on L-form bacteria is continuing. For example, L-form organisms have been observed in mouse lungs after experimental inoculation with Nocardia caviae,[13][14] and a recent study suggested that these organisms may infect immunosuppressed patients having undergone bone marrow transplants.[15] The formation of strains of bacteria lacking cell walls has also been proposed to be important in the acquisition of bacterial antibiotic resistance.[16][17]
L-form bacteria may be useful in research on early forms of life, and in biotechnology. These strains are being examined for possible uses in biotechnology as host strains for recombinant protein production.[18][19][20] Here, the absence of a cell wall can allow production of large amounts of secreted proteins that would otherwise accumulate in the periplasmic space of bacteria.[21][22]
See also[edit]
- Mycoplasmataceae—lack peptidoglycan but supplement their membranes with sterols for stability.
- Protoplast
- Spheroplast
- Ultramicrobacteria
Mycoplasmataceae is a family of bacteria[1] in the order Mycoplasmatales. This family consists of the genera Mycoplasma and Ureaplasma.
In 1967, the order Mycoplasmatales was incorporated into the class Mollicutes.[2] Many species are sexually transmitted and cause pelvic inflammatory disease.[3]
Mycoplasma refers to a genus of bacteria that lack a cell wall and possess a three-layered cellular membrane.[4] They can be parasitic or saprotrophic. Several species are sexually transmitted and pathogenic in humans. Others are found on cats, dogs, and barnyard fowl.
Ureaplasma spp. as human pathogens[edit]
Both Ureaplasma urealyticum and Ureaplasma parvum have been identified as important human pathogens, causing infection in the urogenital tract and, rarely, at distal sites.[5][6][7] Their role in neonatal disease and adverse pregnancy outcomes has been well established, and semantic classifications are changing to reflect the nature of the detrimental outcomes these infections are associated with.[8] In the 2010s, Mycoplasma genitalium has been re-classified as an STI, and it is likely that with more research, Ureaplasma spp. will follow this trend.[9] Similar to other pathogens such as Chlamydia trachomatis, infection with Ureaplasma spp. is associated with adverse fertility outcomes in both men and women.[10][11][12][13][14]Both cause non-gonococcal urethritis. Ureaplasma spp. were implicated in conditions such as prostatitis and chronic pelvic pain syndrome as early as the 1980s.[15][16][17] Research in women has lagged several decades behind, but it is now becoming more clear how Ureaplasma spp. contribute to etiologies such as interstitial cystitis/painful bladder syndrome.[18][19][20] Ureaplasma spp. are associated with alterations in host environment that increase susceptibility to other infections such as bacterial vaginosis and vaginal candidiasis.[21][22] Ureaplasma spp. can cause reactive arthritis as well as directly infect the synovium.[23][7] Some case studies have suggested a causative role in complex regional pain syndrome/reflex sympathetic dystrophy syndrome.[24]
| Mycoplasmataceae | |
|---|---|
| Scientific classification | |
| Kingdom: | |
| Phylum: | |
| Class: | |
| Order: | Mycoplasmatales |
| Family: | Mycoplasmataceae Freundt 1955 |
| Genera | |
Candidatus Hepatoplasma | |
https://en.wikipedia.org/wiki/Mycoplasmataceae
Mollicutes is a class of bacteria[2] distinguished by the absence of a cell wall. The word "Mollicutes" is derived from the Latin mollis (meaning "soft" or "pliable"), and cutis (meaning "skin"). Individuals are very small, typically only 0.2–0.3 μm (200-300 nm) in size and have a very small genome size. They vary in form, although most have sterols that make the cell membrane somewhat more rigid. Many are able to move about through gliding, but members of the genus Spiroplasma are helical and move by twisting. The best-known genus in the Mollicutes is Mycoplasma.
Mollicutes are parasites of various animals and plants, living on or in the host's cells. Many cause diseases in humans, attaching to cells in the respiratory or urogenital tracts, particularly species of Mycoplasma and Ureaplasma. Phytoplasma and Spiroplasma are plant pathogens associated with insect vectors.
Whereas formerly the trivial name "mycoplasma" has commonly denoted any member of the class Mollicutes, it now refers exclusively to a member of the genus Mycoplasma.
| Mollicutes | |
|---|---|
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Tenericutes |
| Class: | Mollicutes Edward and Freundt 1967[1] |
| Orders | |
Acholeplasmatales | |
https://en.wikipedia.org/wiki/Mollicutes
Tenericutes (tener cutis: soft skin) is a phylum of bacteria that contains the class Mollicutes. The name was validated in 1984 as a new division (phylum).[3][4][5] Notable genera include Mycoplasma, Spiroplasma, Ureaplasma, and Phytoplasma.
https://en.wikipedia.org/wiki/Tenericutes
At first, all members of the class Mollicutes were generally named "mycoplasma" or pleuropneumonia-like organism (PPLO). Mollicutes other than some members of genus Mycoplasma were still unidentified. The first species of Mycoplasma/Mollicutes, that could be isolated was Mycoplasma mycoides. This bacterium was cultivated by Nocard and Roux in 1898.[4]
In 1962, R.G.E. Murray proposed to divide the kingdom Bacteria into three divisions (= phyla) on the basis of the cell wall types:
- Gram-negative Gracilicutes, with a thin cell wall and little peptidoglycan;
- Gram-positive "Firmacutes", with a thicker cell wall and more peptidoglycan (the name was later changed in "Firmicutes"), and
- the "Mollicutes", without a cell wall.[6]
https://en.wikipedia.org/wiki/Mollicutes
https://en.wikipedia.org/wiki/Planctomycetes
https://en.wikipedia.org/wiki/Verrucomicrobia
https://en.wikipedia.org/wiki/Chlamydiae
https://en.wikipedia.org/wiki/Planctobacteria
https://en.wikipedia.org/wiki/Aquificae
https://en.wikipedia.org/wiki/Phagemid
https://en.wikipedia.org/wiki/Spumaretrovirinae
https://en.wikipedia.org/wiki/Bovine_leukemia_virus
https://en.wikipedia.org/wiki/Rhizidiovirus
https://en.wikipedia.org/wiki/Plasmaviridae
https://en.wikipedia.org/wiki/Polydnavirus
https://en.wikipedia.org/wiki/Finnlakeviridae
https://en.wikipedia.org/wiki/Mollicutes
https://en.wikipedia.org/wiki/Mycoplasmataceae



No comments:
Post a Comment